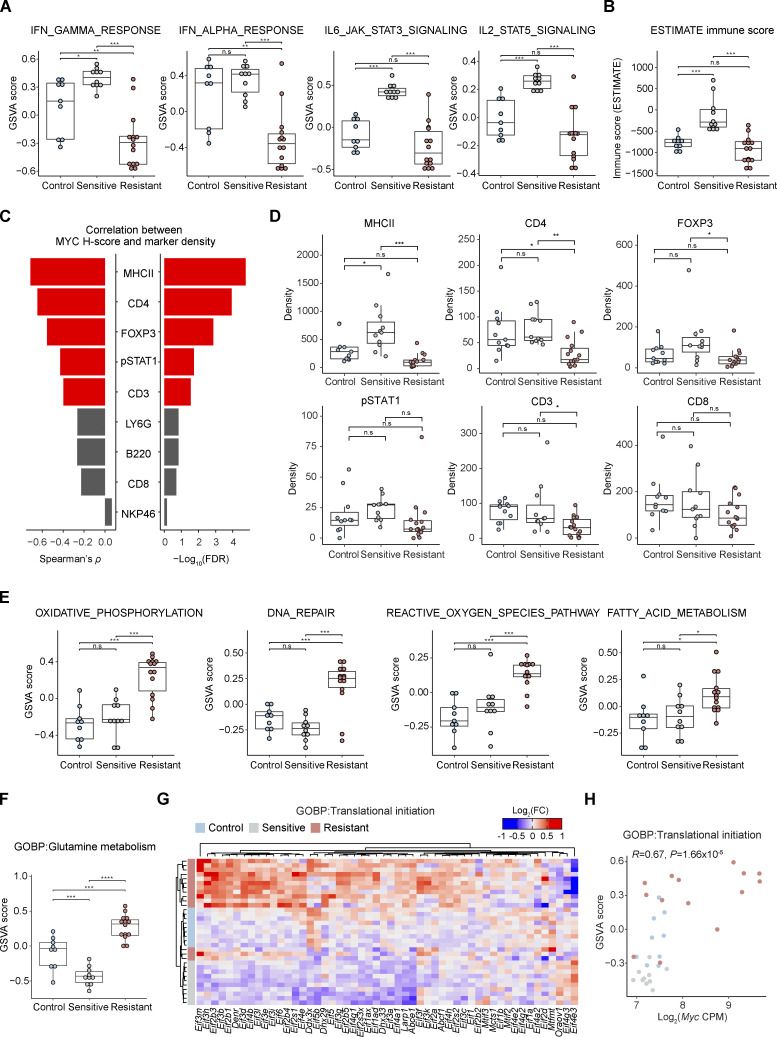
Figure 3.
Biological processes associated with AZD8055 efficacy. (A) GSVA scores for immune-related MSigDB Hallmark gene sets using KEP tumor RNA-seq profiles. (B) Immune infiltration scores based on RNA-seq data inferred by ESTIMATE (Yoshihara et al., 2013). (C) Correlations between MYC protein H-scores and immune cell densities based on quantified IHC for MYC and indicated immune markers. Spearman’s ρ and −log10(FDR) are depicted as bar plots and red bars indicate significant correlations (FDR < 0.05). P values were calculated with two-tailed t-transformations of Spearman’s correlation coefficient. (D) Immune cell densities based on quantified IHC for indicated immune markers. (E and F) GSVA scores for MSigDB Hallmark gene sets (E) and GO glutamine metabolism (F) upregulated in resistant tumors. (G) Heatmap displaying mRNA expression of translation initiation genes obtained from the GO Biological Processes (GOBP) translational initiation gene set. Hierarchical clustering (Euclidean distance and complete-linkage clustering) was performed on log2-fold-change (log2FC) of CPM values centered on the median expression of control tumors. (H) Pearson’s correlation analysis of Myc mRNA expression (log2CPM) vs. GOBP translation initiation gene set scores. P value was calculated with two-tailed t-transformations of Pearson’s correlation coefficient. In A, B, and E–H, 9 control, 10 sensitive, and 14 resistant KEP tumors, and in C and D, 13 control, 11 sensitive, and 15 resistant KEP tumors were analyzed. In A, B, and D–F, data are represented as median ± IQR (box) and quartiles ± 1.5 × IQR (whiskers), and one-way ANOVA and Tukey’s post-hoc tests were performed to compute adjusted P values (*P < 0.05; **P < 0.01; ***P < 0.001; n.s, not significant).