FIGURE 1.
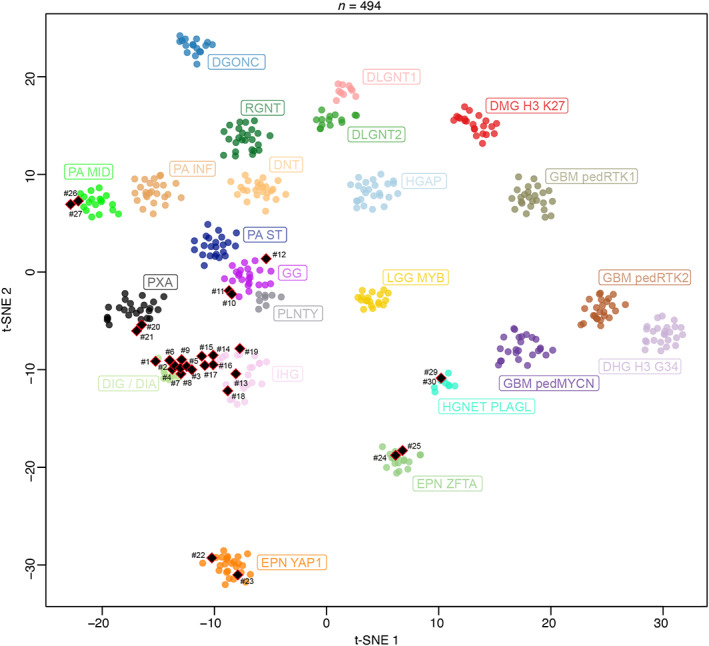
DNA methylation‐based t‐distributed stochastic neighbor embedding distribution. Our tumors were compared to the following reference DNA methylation classes. Diffuse glioneuronal tumor with oligodendroglioma‐like features and nuclear clusters (DGONC), diffuse hemispheric glioma, H3.3 G34 mutant (DHG H3 G34), desmoplastic infantile ganglioglioma/desmoplastic infantile astrocytoma (DIG/DIA), diffuse leptomeningeal glioneuronal tumor, subtype 1 (DLGNT1), diffuse leptomeningeal glioneuronal tumor, subtype 2 (DLGNT2), diffuse midline glioma H3 K27M mutant (DMG H3K27), dysembryoplastic neuroepithelial tumor (DNT), supratentorial ependymoma, YAP1 fusion‐positive (EPN YAP1), supratentorial ependymoma, ZFTA fusion‐positive (EPN ZFTA), pediatric glioblastoma, IDH wildtype, subclass MYCN (GBM pedMYCN); pediatric glioblastoma, IDH wildtype, subclass RTK1 (GBM pedRTK1), pediatric glioblastoma, IDH wildtype, subclass RTK2 (GBM pedRTK2), ganglioglioma (GG), high‐grade astrocytoma with piloid features (HGAP), high‐grade neuroepithelial tumor, with PLAG‐family amplification (HGNET PLAGL), infant‐type hemispheric glioma (IHG), low‐grade glioma, MYB (LGG MYB), infratentorial pilocytic astrocytoma (PA INF), midline pilocytic astrocytoma (PA MID), supratentorial pilocytic astrocytoma (PA ST), polymorphous low‐grade neuroepithelial tumor of the young (PLNTY), pleomorphic xanthoastrocytoma (PXA), and rosette forming glioneuronal tumor (RGNT).
