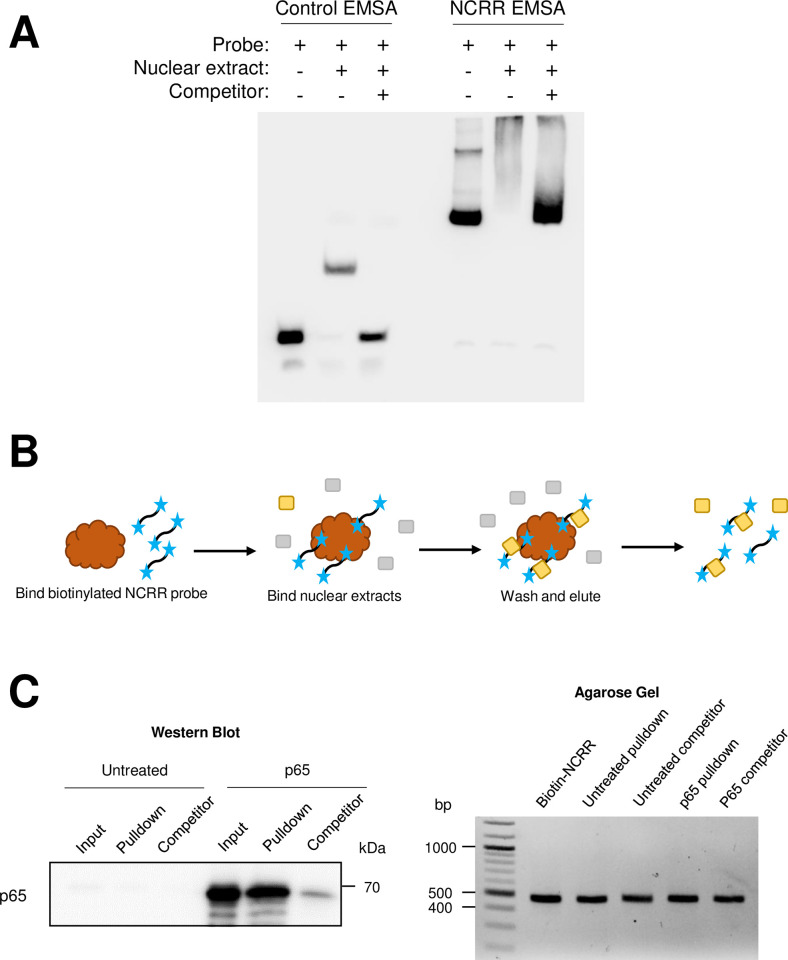
Fig 6. NF-κB p65 binds directly to the MCPyV NCRR.
(A) NCRR-specific DNA binding activity is detected in HEK293 nuclear extracts containing overexpressed p65. EMSA was performed using a set of positive control probes and nuclear extract provided in the LightShift Chemiluminescent EMSA kit (“Control EMSA”) or using full NCRR probes and nuclear extracts from HEK293 cells transfected with a p65-expressing plasmid (“NCRR EMSA”). (B) Schematic of the biotinylated DNA pulldown assay. Biotinylated (blue stars) NCRR probes were bound to streptavidin-coated magnetic beads (brown), then incubated with nuclear extracts containing the protein of interest (in yellow; other nuclear proteins are indicated in gray). Alternatively, the nuclear extracts are pre-incubated with an excess amount of unlabeled NCRR probe before being incubated with the bead-bound probes. Protein-probe complexes (protein of interest [yellow] bound to biotinylated [blue stars] probes) are eluted off the beads for analysis by SDS-PAGE/Western blot or agarose gel electrophoresis. (C) NF-κB p65 binds the MCPyV NCRR. Biotinylated NCRR pulldown assays were performed with nuclear extracts from untreated HEK293 cells (“Untreated”), or cells overexpressing p65 (“p65”), and biotinylated-NCRR probes in the presence or absence of an excess of unlabeled NCRR competitor (“Competitor”). The left panel depicts the detection of p65 by Western blotting in the input (1%) and pulldown samples, while the right panel demonstrates that comparable amounts of biotinylated probe were bound to the beads in each pulldown experiment.