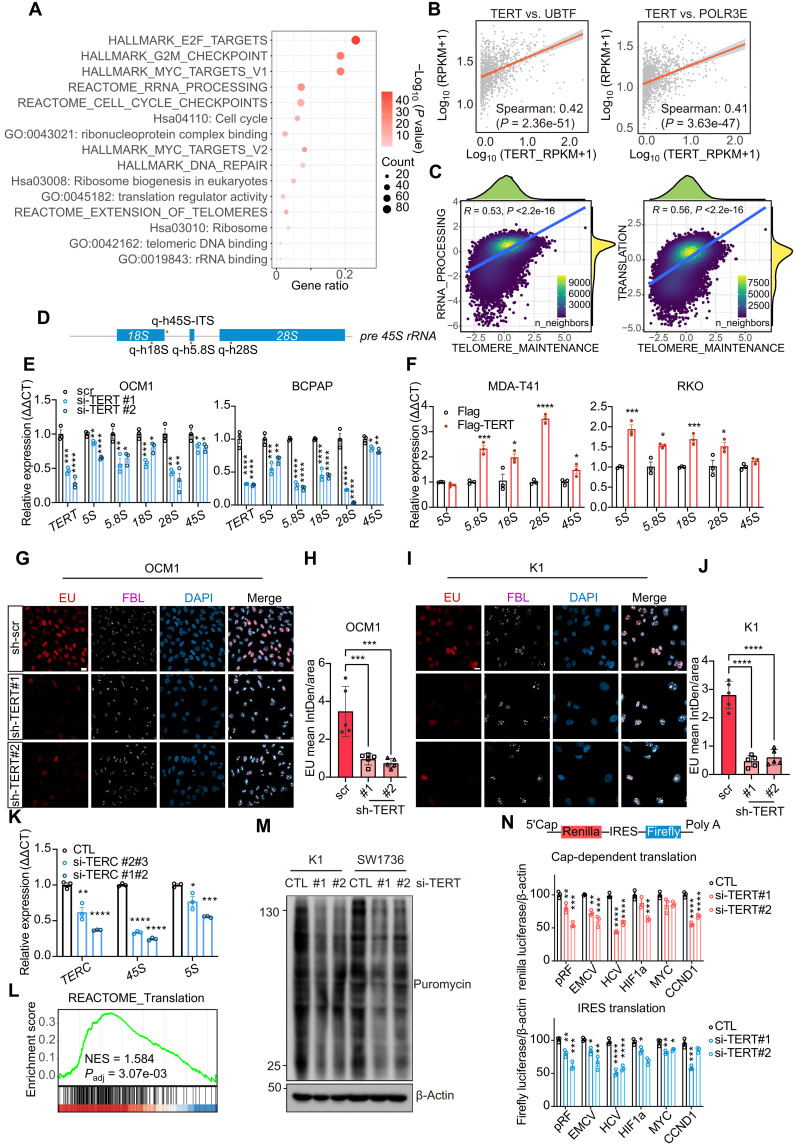
Fig. 4. TERT induces rRNA synthesis and ribosomal activity.
(A) Enrichment results of TERT-positive correlation genes (R > 0.25) in CCLE database (n = 1156). (B) Correlation between TERT and UBTF, POLR3E (n = 1156). (C) Correlation of Reactome: telomere maintenance and rRNA processing/translation enrichment scores in THCA single-cell data of thyroid cells (n = 42,708 cells). (D) Schematic diagram of pre-45S rRNA and the positions of qPCR primers. (E and F) qPCR results of rRNA 5S, 5.8S, 18S, 28S, and 45S change after TERT knockdown [(D); in OCM1 and BCPAP cells] and TERT overexpression [(E); in MDA-T41 and RKO cells]. Data represent means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test in (D) and Student’s t test in (E). (G and I) EU (red) staining and FBL (white) immunofluorescence results after TERT knockdown. Scale bar, 20 μm. (H and J) Quantification results of (G) and (I). EU staining results were quantified using ImageJ software (n = 5). Data represent means ± SD. ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test. (K) qPCR results of TERC, 45S, and 5S after TERC knockdown in BCPAP cells. Data represent means ± SEM (n = 3). (L) GSEA result of between TERT high-expression (n = 538) and low-expression (n = 538) cells in CCLE datasets. (M) SUnSET assay results after TERT siRNA treatment of K1 and SW1736. (N) pRF, EMCV, HCV, HIF-1a, MYC, and CCND1 bicistronic reporter assay results of HEK293T after TERT siRNA treatment. The ratios represented the relative luciferase reads normalized by separative β-actin protein levels. Data represent means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test.