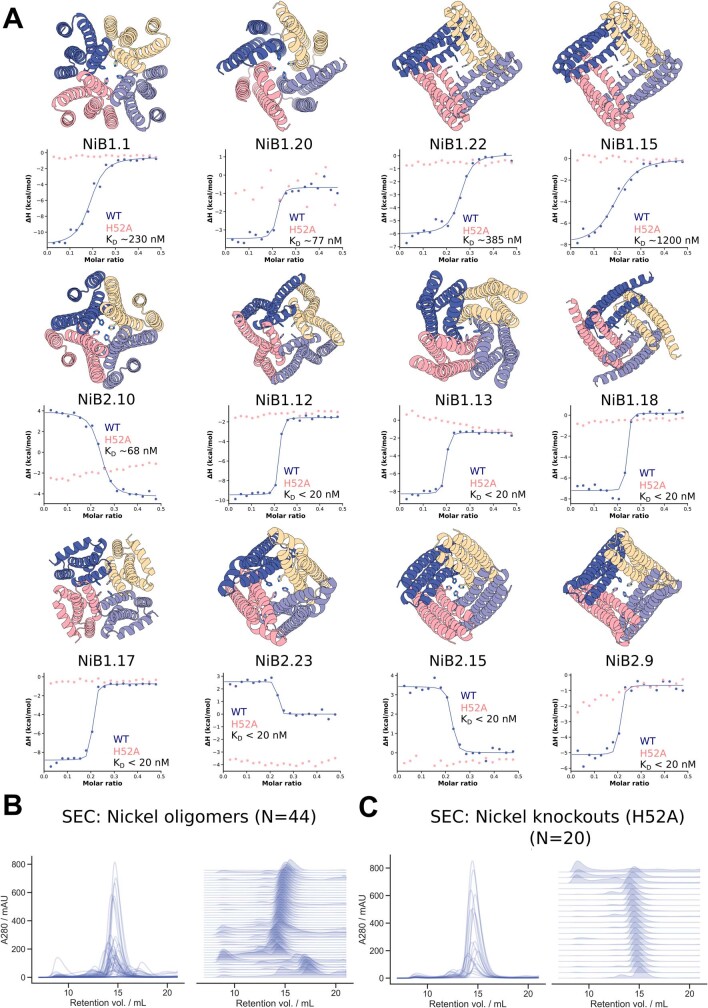
Extended Data Fig. 7. Additional Ni2+ binding C4 oligomers.
A) AF2 predictions of a subset of the experimentally verified Ni2+ binding oligomers, with corresponding isothermal titration calorimetry (ITC) binding isotherms for the wild-type (blue) and H52A mutant (pink) below. Note that these, with Fig. 5, encompass all of the experimentally validated outputs deriving from unique RFdiffusion backbones. Wild-type dissociation constants are displayed in each plot. We observe a mixture of endothermic (NiB2.10, NiB2.23, NiB2.15) and exothermic isotherms. For all cases displayed we observe no binding to the ion for H52A mutants, indicating the scaffolded histidine at position 52 is critical for ion binding. KD values in the isotherms indicate binding of the ion with the designed stoichiometry (1:4 Ni2+:protein). Note that each backbone depicted is from a unique RFdiffusion sampling trajectory, and that models and data for designs NiB2.15, NiB1.12, NiB1.20 and NiB1.17 from Fig. 5 are duplicated here for ease of viewing. B) Size exclusion chromatograms for elutions from the 44 purifications suggest the vast majority of designs are soluble and have the correct oligomeric state. C) Size exclusion chromatograms for 20 H52A mutants show that the mutants remain soluble and retain the intended oligomeric state. Note that only 18 of these 20 had wild-type sequences that definitively bound nickel. Note also that for ITC plots, points represent single measurements.