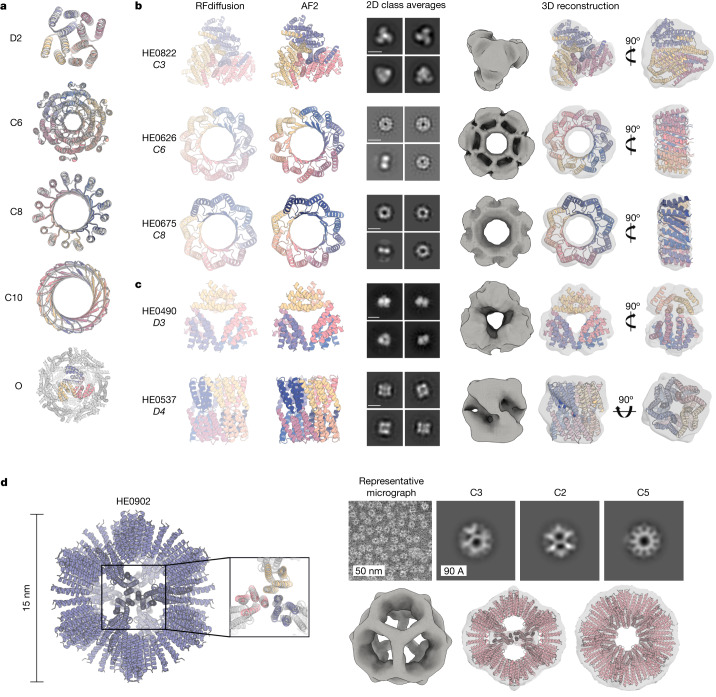
Fig. 3. Design and experimental characterization of symmetric oligomers.
a, RFdiffusion-generated assemblies overlaid with the AF2 structure predictions based on the designed sequences; in all five cases they are nearly indistinguishable (for the octahedron (bottom), the prediction was for the C3 substructure). Symmetries are indicated to the left of the design models. b,c, Designed assemblies characterized by nsEM. Model symmetries are as follows: cyclic, C3 (HE0822, 350 amino acids (AA) per chain), C6 (HE0626, 100 AA per chain) and C8 (HE0675, 60 AA per chain) (b); dihedral, D3 (HE0490, 80 AA per chain) and D4 (HE0537, 100 AA per chain) (c). From left to right: (1) symmetric design model, (2) AF2 prediction of design following sequence design with ProteinMPNN, (3) 2D class averages showing both top and side views (scale bar, 60 Å for all class averages) and (4) 3D reconstructions from class averages with the design model fit into the density map. The overall shapes are consistent with the design models, and confirm the intended oligomeric state. As in a, AF2 predictions of each design are nearly indistinguishable from the design model (backbone r.m.s.d.s (Å) for HE0822, HE0626, HE0490, HE0675 and HE0537, are 1.33, 1.03, 0.60, 0.74 and 0.75, respectively). d, nsEM characterization of an icosahedral particle (HE0902, 100 AA per chain). The design model, including the AF2 prediction of the C3 subunit are shown on the left. nsEM data are shown on the right: on top, a representative micrograph is shown alongside 2D class averages along each symmetry axis (C3, C2 and C5, from left to right) with the corresponding 3D reconstruction map views shown directly below overlaid on the design model.