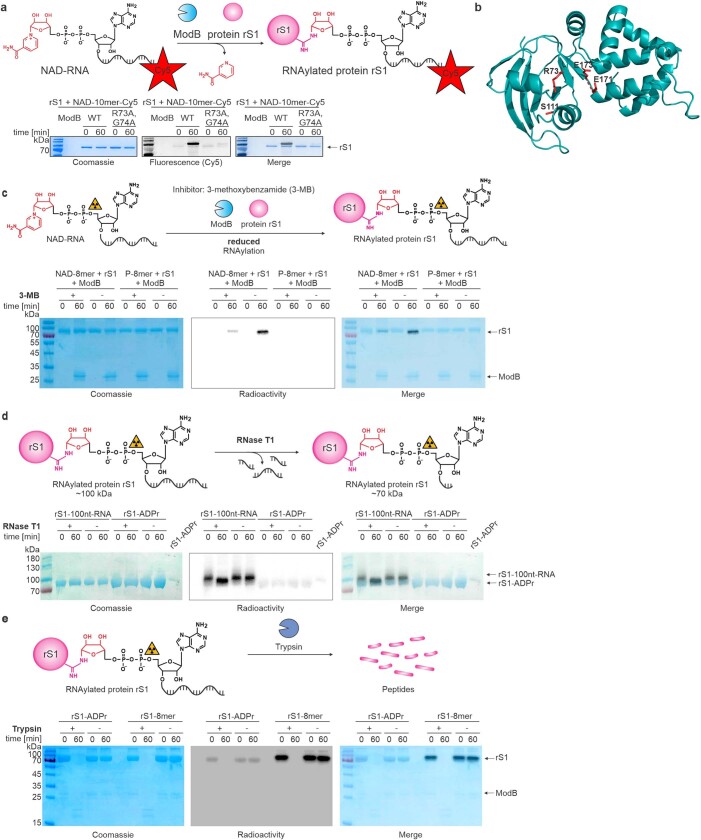
Extended Data Fig. 2. Characterisation of the RNAylation of protein rS1 by ModB.
a, RNAylation of rS1 in the presence of catalytically active ModB and catalytically inactive ModB R73A, G74A with NAD-10mer-Cy5 (n = 3). In addition to the catalytically important residue R73, we mutated G74 as well. Mutation of G74 results in an altered PAM region, which is important for CRISPR-Cas9 gene editing of the T4 phage genome. b, AlphaFold prediction63 of the structure of ModB. Active site residues of the R-S-EXE motif1 are highlighted in red. Corresponding confidence metrics are shown in Supplementary Fig. 2. c, Inhibition of in vitro RNAylation of protein rS1 by ModB via ART inhibitor 3-MB. Reactions were performed with 32P-NAD-RNA 8mer (32P-NAD-8mer) as well as 32P-RNA 8mer (negative control) (n = 3). 3-MB reduces the yield of RNAylated rS1. d, in vitro digest of RNAylated and ADP-ribosylated protein rS1 by RNase T1. Reactions performed in the absence of RNase T1 (-) serve as negative controls. Protein rS1 ADP-ribosylated in the presence of 32P-NAD was applied as a reference (S1-ADPr) (n = 2). Upon T1 digest, the 100nt-RNA at rS1 is shortened, and the molecular weight of RNAylated rS1 is reduced. This leads to similar electrophoretic mobility as for ADP-ribosylated rS1. e, Tryptic digest of ADP-ribosylated and RNAylated protein rS1 (n = 2). The protein is degraded in the presence of trypsin, and RNAylation and ADP-ribosylation signals are lost. All samples were analysed by 12 % SDS-PAGE, protein was visualised by Coomassie staining and RNAylation was assessed via a radioactivity scan.