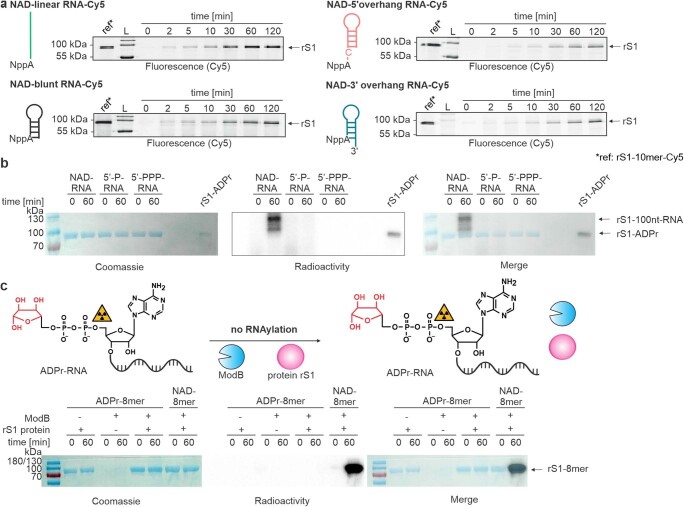
Extended Data Fig. 3. Characterisation of ModB mediated RNAylation in vitro.
a, Analysis of the role of RNA secondary structure on RNAylation reaction. Four different 3′-Cy5-labelled NAD (NppA)-capped RNAs were tested including a linear (green) NAD-capped RNA (10mer) and three structured RNAs with either a 3′-overhang (blue), a 5′-overhang (red) or a blunt end (black) (n = 3). SDS-PAGE analyses of the time course of RNAylation are shown. Quantification of the signal intensities (Cy5 scan) relative to the reference is shown in Fig. 2c. ModB prefers linear 5′-ends of NAD-capped RNAs. L = ladder. b, Analysis of the RNAylation dependency on the presence of a 5′-NAD-cap of the RNA. 10 % SDS-PAGE analysis of in vitro RNAylation of the protein rS1 by ModB in the presence of either 5′-NAD-capped (NAD-RNA), 5′-monophosphate- (5′-P-RNA) or 5′-triphosphate-100nt-RNA (5′-PPP-RNA) (n = 2). RNAylation with radiolabelled RNA is detected by radioactivity scan and protein load visualised by Coomassie staining. In vitro RNAylation of rS1 is only observed in the presence of NAD-RNA. RNAylated rS1 cannot be detected by Coomassie staining due to low sensitivity. c, Characterisation of ADPr-RNA (which is lacking the nicotinamide moiety compared to NAD-RNA) as a substrate for ModB (n = 2). As a positive control, NAD-8mer was applied. All reactions were analysed by 12 % SDS-PAGE. RNAylation with radiolabelled RNA is detected by radioactivity scan and protein load visualised by Coomassie staining. ADPr-RNA is not accepted as a substrate for ModB-catalysed RNAylation in vitro.