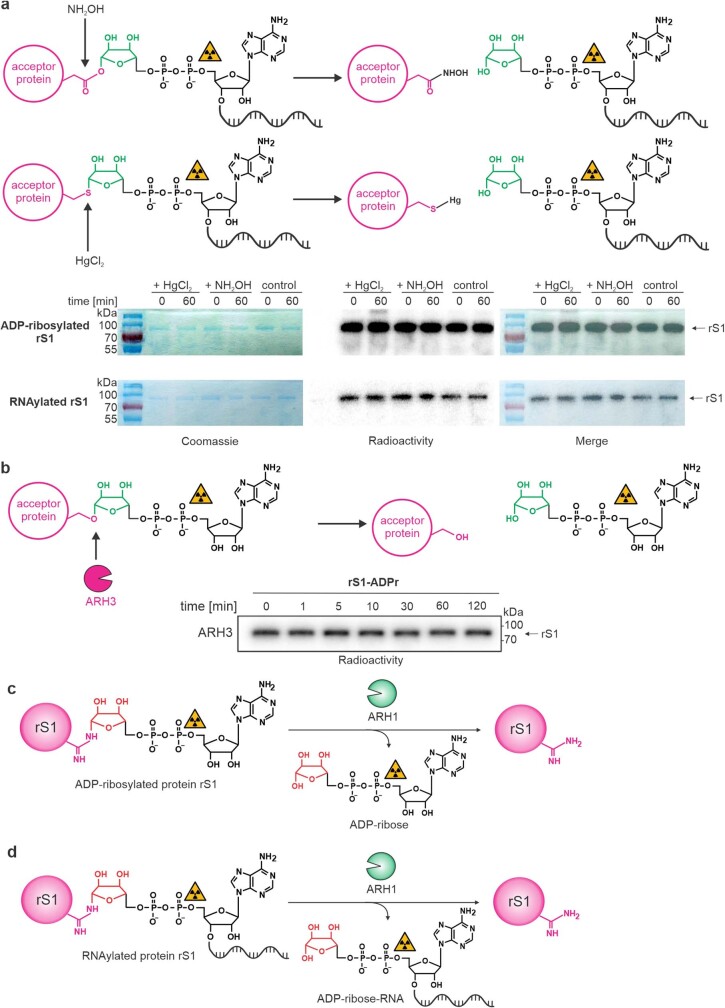
Extended Data Fig. 4. Specific removal of RNAylation using chemical and enzymatic treatments.
a, Different ADP-ribose-protein linkages have been shown to be either stable or unstable in the presence of HgCl2 and neutral hydroxylamine (NH2OH), which represents a relatively straightforward and fast approach to identify ADP-ribosylation sites. Treatment with NH2OH hydrolyses linkages between glutamate/aspartate and ADP-ribose. HgCl2 specifically cleaves thiol-glycosidic bonds. ADP-ribosylated and RNAylated protein rS1 were treated with NH2OH or HgCl2. The removal of ADPr or RNA by these chemicals would result in a decrease of the radioactive signal of protein rS1. All samples were analysed by 12 % SDS-PAGE, stained in Coomassie (protein loading control) and RNAylation assessed as radioactivity. A decrease of the radioactive signal in comparison to the control (untreated) was not determined (n = 1). b, in vitro time course of the stability of rS1 ADP-ribosylation in the presence of ARH3 analysed by 12 % SDS-PAGE (n = 1). The autoradiography scan is presented. ARH3 did not remove the ADP-ribosylation. c-d, Reaction schematics for the removal of the ADP-ribosylation (c) and RNAylation (d) of rS1 by ARH1.