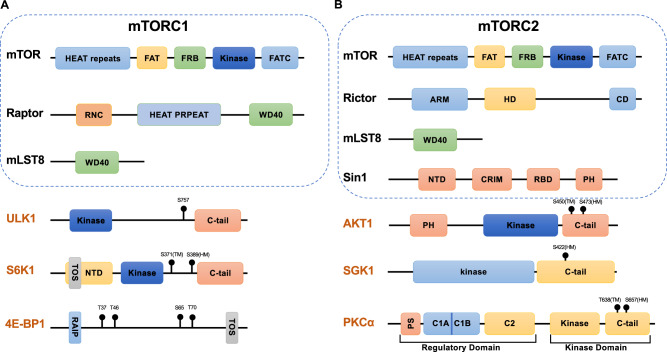
Fig. 3.
Composition of mTORC1/2 core subunits and selected substrates. A Main protein domains and phosphorylation sites of mTORC1 core subunits and selected substrates. mTOR and mLST8 are shared subunits of mTORC1/2, while Raptor and Rictor/Sin1 are the defining subunits for mTORC1 and mTORC2, respectively. S6K is phosphorylated by mTORC1 at S371 (TM) and T389 (HM) in the linker region. 4E-BP utilizes the TOS and RAIP motif to interact with Raptor/mTORC1 and is sequentially phosphorylated by mTORC1 at T37/T46 and S65/T70. ULK1 can be phosphorylated on S757. B Main protein domains and phosphorylation sites of mTORC2 core subunits and selected substrates. MLST8 interacts with Sin1 to position its substrate-interacting CRIM domain, providing substrate specificity of mTORC2. Sin1/mTORC2 phosphorylates T450 (turn motif) and S473 (hydrophobic motif) in the C-tail of Akt1. This dual phosphorylation has also been observed in PKC, while for SGK1, S422(HM) is the only known site phosphorylated by mTORC2. HEAT repeat found in Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1, FAT FAK focal adhesion targeting, FRB FKBP-rapamycin-binding, FATC FRAP-ATM-TRRAP-C-terminal, RNC Raptor N-terminal conserved, NTD N-terminal domain, TOS TOR signaling, RAIP Arg-Ala-Ile-Pro motif, ARM Armadillo, HD HEAT-like domain, CD C-terminal domain, CRIM conserved region in the middle, RBD Ras-binding domain, PH Pleckstrin homology, PS pseudosubstrate, C1,C2 membrane targeting module