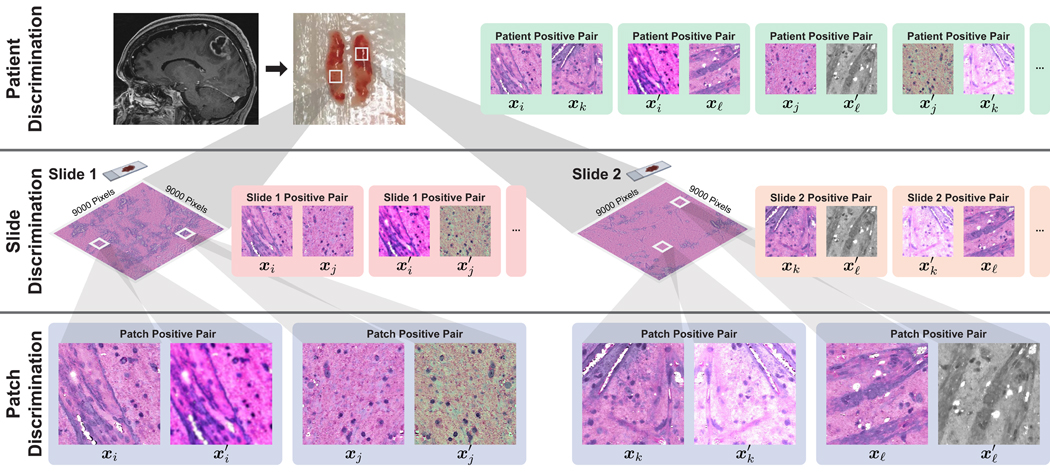
Figure 2. HiDisc Overview.
Motivated by the patient-slide-patch data hierarchy of clinical biomedical microscopy, HiDisc defines a patient, slide, and patch discriminative learning objective to improve visual representations. Because WSI and microscopy data are inherently hierarchical, defining a unified hierarchical loss function does not require additional annotations or supervision. Positive patch pairs are defined based on a common ancestry in the data hierarchy. A major advantage of HiDisc is the ability to define positive pairs without the need to sample from or learn a set of strong image augmentations, such as random erasing, shears, color inversion, etc. Because each field-of-view in a WSI is a different view of a patient’s underlying cancer diagnosis, HiDisc implicitly learns image features that predict that diagnosis.