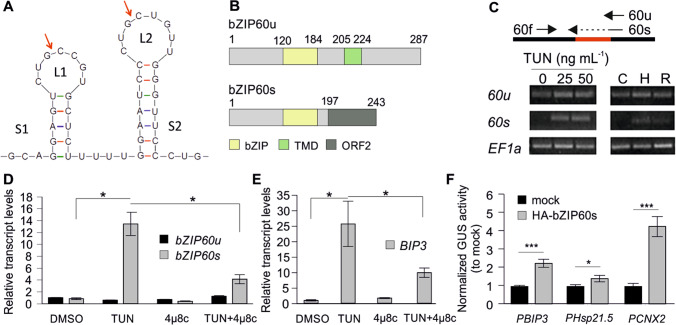
Fig. 1.
Tomato bZIP60 regulation and transactivation activity. A Double stem (S)-loop (L) structure of the bZIP60u mRNA recognized by IRE1 as predicted by RNAFold (Zuker and Stiegler 1981). Arrows indicate the cleavage sites. B Domain structure of bZIP60u and bZIP60s. Numbers indicate amino acid positions. bZIP, basic leucine zipper; TMD, transmembrane domain; ORF2, open reading frame generated by the frame shift. C RT-PCR of bZIP60u (unspliced) using 60f/60u oligonucleotides and bZIP60s (spliced) using 60f/60 s oligonucleotides, in seedlings exposed to DMSO (0 ng mL−1), 25 or 50 ng mL−1 of TUN. Seedlings were also exposed to a heat stress regime: 25 °C as control (C), 40 °C for 1 h (H), and then allowed to recover at 25 °C for 1.5 h (R). EF1a is the housekeeping gene. Relative transcript levels of D bZIP60u and bZIP60s or E BIP3 in young tomato leaves treated with DMSO or TUN (50 ng mL−1) and/or 4μ8c (0.25 µM). Relative transcript levels were determined by qRT-PCR using 2−ΔΔ.Ct method, EF1a as reference gene and normalized against DMSO WT as control sample. Each value is the average of three independent biological replicates. Error bars are ± SEM and asterisks depict significant differences based on ANOVA with Duncan’s multiple range test (*p < 0.05) for each gene. F Transactivation activity of bZIP60s using GUS reporter assay on promoters (1 kb) of selected tomato genes. Values are average of normalized GUS activity relative to mock control (n = 3). Error bars are ± SEM and asterisks indicate significant differences based on Student’s t-test (*p < 0.05, ***p < 0.001)