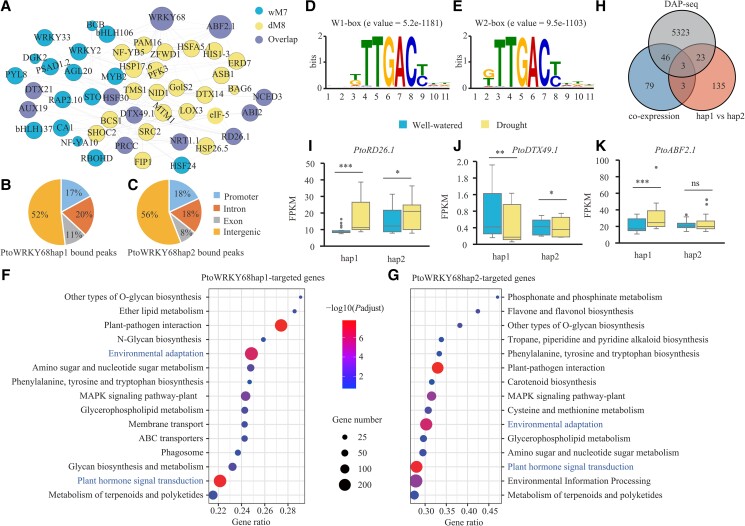
Figure 3.
Identification of genome-wide direct targets of PtoWRKY68hap1 and PtoWRKY68hap2. A) Genes identified in the well-watered network module 7 (wM7), the drought-stressed module 8 (dM8) (containing PtoWRKY68) and overlapping genes under two conditions (Overlap) with putative Arabidopsis orthologs are shown in network view. Node size indicates module membership. The larger nodes are highly connected within a module. B and C) Whole-genome distribution of bound genes by PtoWRKY68hap1B) and PtoWRKY68hap2C) obtained by DAP-seq with P < 0.05. Promoter regions were defined as the binding peaks within 2 kb upstream of ATG. D and E) Binding motifs of PtoWRKY68hap1 (D, W1-box) and PtoWRKY68hap2 (E, W2-box) protein by MEME-ChIP. F and G) The top KEGG-enriched terms of PtoWRKY68hap1F) and PtoWRKY68hap2G) bound genes by DAP-seq. P-value by Fisher's test. H) Venn diagram showing that three overlapping genes were considered PtoWRKY68 alleles direct genes, were co-expressed with PtoWRKY68, were differentially expressed in PtoWRKY68hap1 vs. PtoWRKY68hap2, and were directly bound by PtoWRKY68 alleles. I to K) FPKM values of PtoRD26.1I), PtoDTX49.1J), and PtoABF2.1K) in hap1 and hap2 haplogroup under well-watered and drought-stress conditions. In box plots, center line represents the median, box limits denote the upper and lower quartiles, whickers indicate the interquartile range, and dots are outliers. Significant differences were determined using the t-test, *P < 0.05, **P < 0.01, ***P < 0.001. ns, no significant difference.