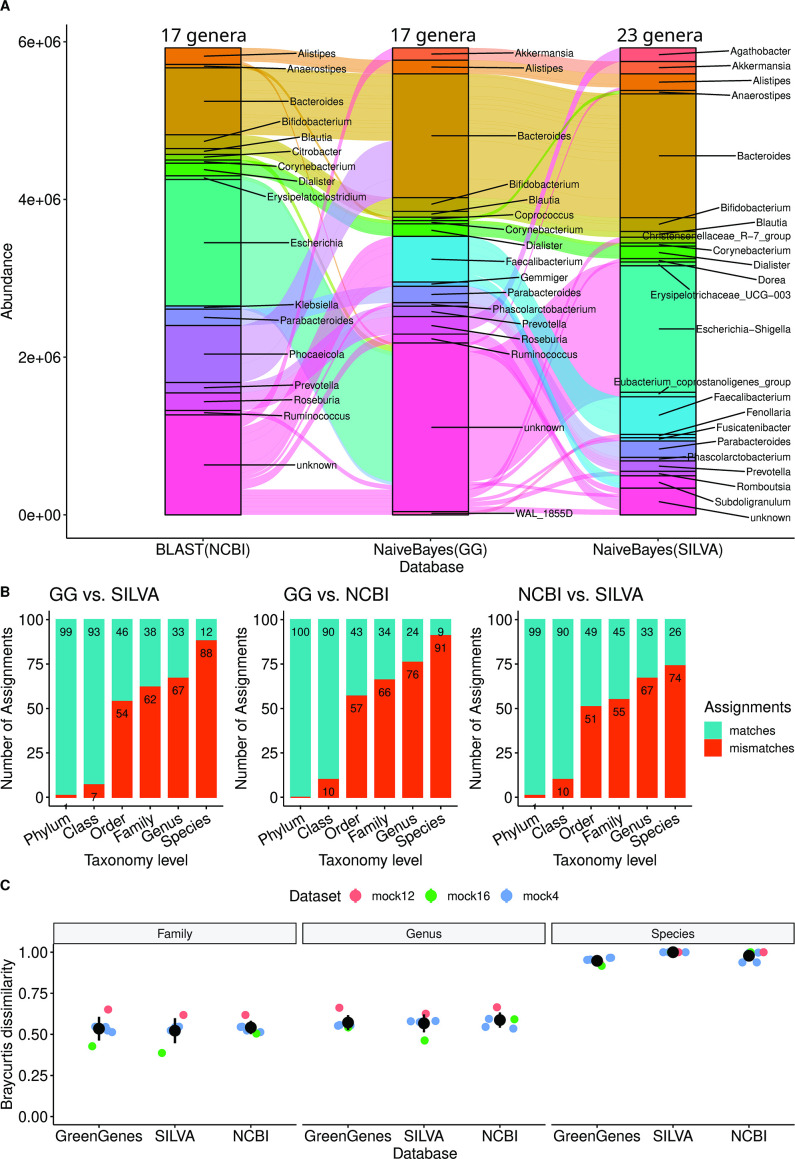
Fig 3.
Taxonomic reference databases vary in terms of their taxonomy assignments below the order level. (A) The taxonomic assignments of the top 50 representative sequences using the three different reference databases. This result illustrates how the same sequences are assigned to different genera under different databases. A significant portion of the representative sequences is assigned to an “unknown” genus in two of three databases (GreenGenes and NCBI). The number of assigned genera for each database is displayed at the top of each column. (B) The number of representative sequences assigned to the same taxonomic label when using different reference databases (for the top 100 representative sequences). The mismatches are fewer at higher taxonomic levels but, even at the order level there exists greater than 51% of mismatches, demonstrating the poor agreement in taxonomic labels assigned by the different databases. The data used for the analysis in (A and B) were samples (healthy and ASD) from the FMT data set. (C) The Bray-Curtis dissimilarity between the predicted taxonomy profile and expected taxonomy profile in the mock data sets shows that there is no singular best choice of database for every data set, as all the databases show similar performances. The GreenGene database and the Naive Bayes classifier are chosen as the defaults for the TA step of MiCoNE due to their popularity. The data sets used for the analysis in (C) were the mock data sets from mockrobiota.