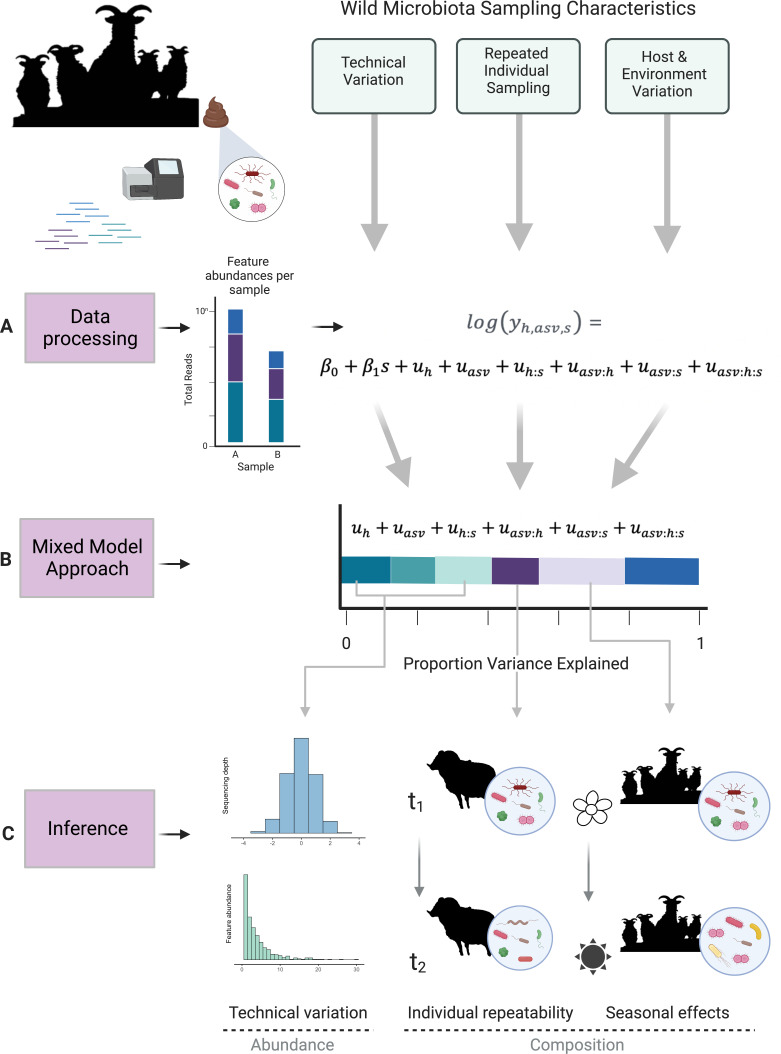
Fig 1.
Overview of mixed-model approach to wild microbiota analysis. Data processing (A) generates amplicon sequence variant (ASV)–level abundances for each sample. These raw abundances are used as the response for generalized linear mixed-effects models with Poisson error families. In the example illustrated, data include sampling time points for a group of individuals taken during two seasons. Model syntax therefore specifies a fixed effect of age, and random effects for taxonomy (asv), sample id (host:season, h:s), individual differential abundance of ASVs (asv:h), differential abundance of ASVs across seasons (asv:s), and a residual variance at the row level (asv:h:s). GLMM output can be used to partition the variance explained by each random-effect term (B). These variance components can be interpreted as the relative contributions of both technical variation and host or environmental contributions to differential abundance as illustrated in (C). Created with BioRender.com.