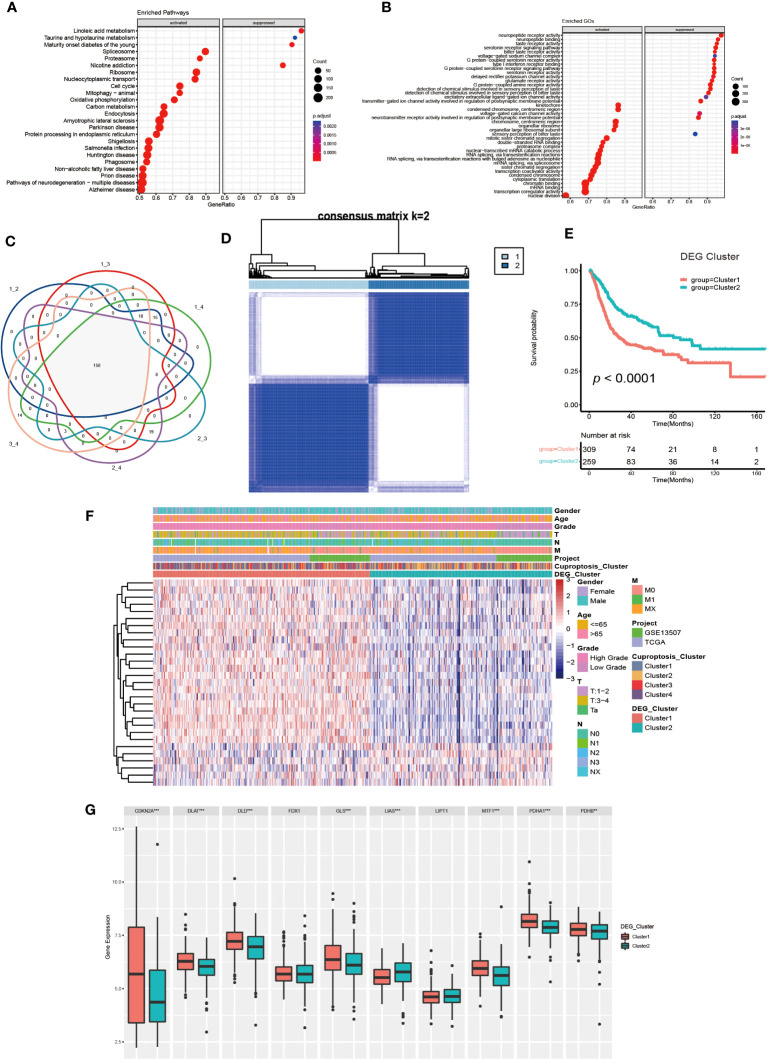
Figure 4.
Generation and functional annotation of differentially expressed genes (DEGs) based on cuproptosis cluster. (A, B) The functional annotation for cuproptosis cluster-related DEGs using KEGG (A) and GO (B) enrichment analysis. (C) Cuproptosis clusters-related DEGs (N=150) shown in the Venn diagram. (D) The unsupervised consensus clustering of cuproptosis clusters-related DEGs (N=150) in the training set and consensus matrices for k =2. (E) The differences in OS between two DEG clusters by Kaplan-Meier curves. (F) The unsupervised consensus clustering analysis to classify patients into two different DEG subtypes and clinical characteristics between the two DEG clusters. (G) The expression of 10 CRGs between two DEG clusters (**p< 0.01, ***p< 0.001).