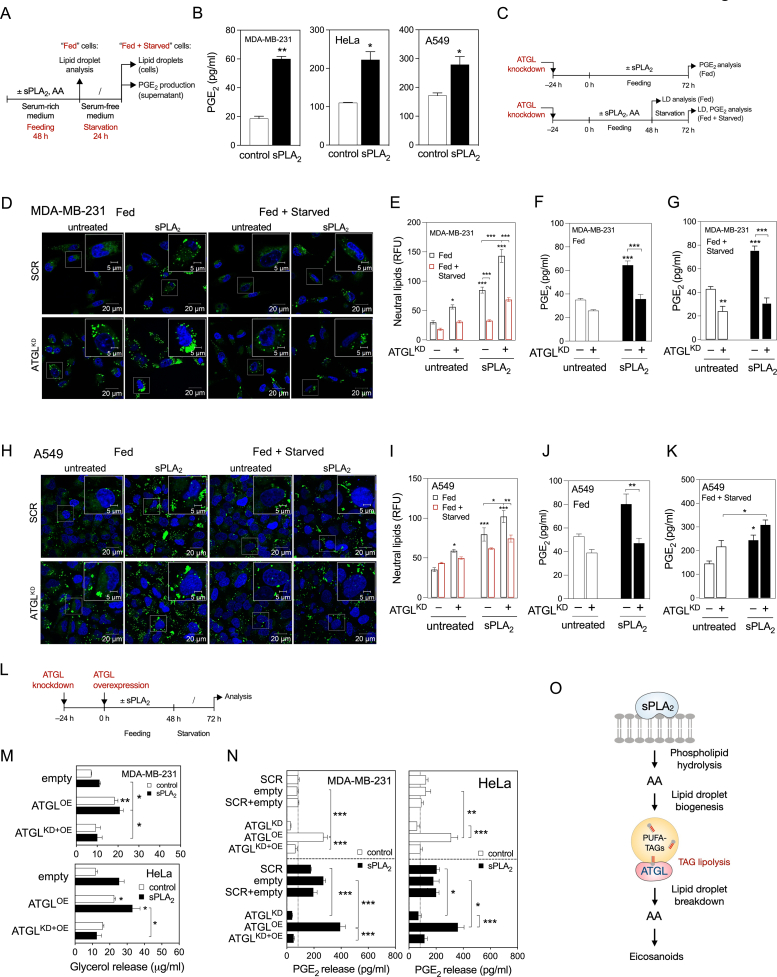
Figure 2.
ATGL-mediated LD breakdown is required for PGE2production. (A) Diagram illustrating the experimental set-up used to load cells with LDs (Feeding) and then to induce their breakdown (Starvation). (B) PGE2 levels in cell supernatants of control and hGX-sPLA2-treated cells quantified by ELISA at the end of the starvation period. (C) Diagram illustrating the experimental conditions used in (D)–(K) and Supp. Figure 2(E)–(L). (D–F, H–J) LD levels and PGE2 production in ATGL-silenced control and sPLA2-treated MDA-MB-231 and A549 cells grown as shown in (C) and analysed after 72 h of feeding (Fed) or after 48 h feeding plus 24 h starvation (Fed + Starved). Neutral lipids were quantified by Nile Red staining and flow cytometry, PGE2 was quantified by ELISA. (G, K) Representative confocal microscopy images showing effects of ATGL depletion on cellular LD content in control and hGX-sPLA2-treated MDA-MB-231 and A549 cells, under serum-rich (Fed) and serum-free (Fed + Starved) conditions. LDs and nuclei were stained using BODIPY 493/503 and Hoechst 33342, respectively. (L) Diagram illustrating the experimental set-up used in (M), (N) and Supp. Figure 2M, N. (M, N) Glycerol release and PGE2 production in ATGL-overexpressing serum-starved cells (both untreated and hGX sPLA2 pretreated), in comparison with cells co-transfected with ATGL-specific siRNAs (ATGLKD + OE), non-targeting siRNA (scrambled) and control plasmid (empty), grown as illustrated in (L). (O) Diagram illustrating the proposed model of LD-mediated eicosanoid production in cancer cells. Data are means ± SEM of two (M) or three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-way ANOVA with Tukey (N) or Bonferroni (E–G, I–K, M) adjustment; unpaired t-tests (B)).