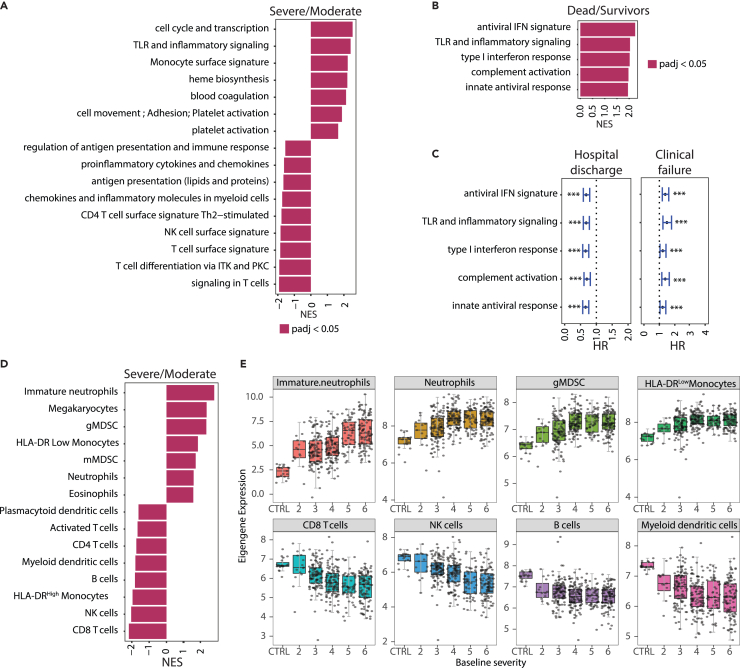
Figure 3.
Transcriptomic signatures associated with COVID-19 severity and disease progression
(A) Fast gene set enrichment analysis (FGSEA) of differentially expressed genes between cases with baseline severity ≥4 (Severe, n = 280) and <4 (Moderate, n = 124) at baseline. Higher NES represents upregulation of the indicated immune pathways.
(B) FGSEA analysis of differentially expressed genes between cases associated with higher mortality (death by day 28, n = 83) and survivors (n = 321). Higher NES represents upregulation of the indicated immune pathway.
(C) Forest plot showing hazard ratios with 95% confidence ratios identified using Cox proportional hazard model depicting time to hospital discharge (left) and clinical failure (right) from COVID-19 for immune pathways shown in (B). ∗∗∗Represents Benjamini-Hochberg adjusted p value < 0.05. Error bars represent 95% confidence intervals.
(D) Same as 3A where higher NES represents upregulation of blood cellularity signatures.
(E) Boxplots showing changes in eigengene values of blood cell-type signatures across different baseline severity scales of COVID-19 patients. The X axis represents the baseline severity scales: 2 (n = 15), 3, (n = 109), 4 (n = 114), 5 (n = 58), 6 (n = 107) and healthy controls (CTRL, n = 19). CD, cluster of differentiation; gMDSC, granulocytic myeloid-derived suppressor cells; HLA, human leukocyte antigen; HR, hazard ratio; IFN, interferon; ITK, IL-2 inducible T cell kinase; mMDSC, monocytic myeloid-derived suppressor cells; NES, normalized enrichment score; NK, natural killer; PKC, protein kinase C; PBO, placebo; TCZ, tocilizumab; Th, T helper cell; TLR, toll-like receptors.