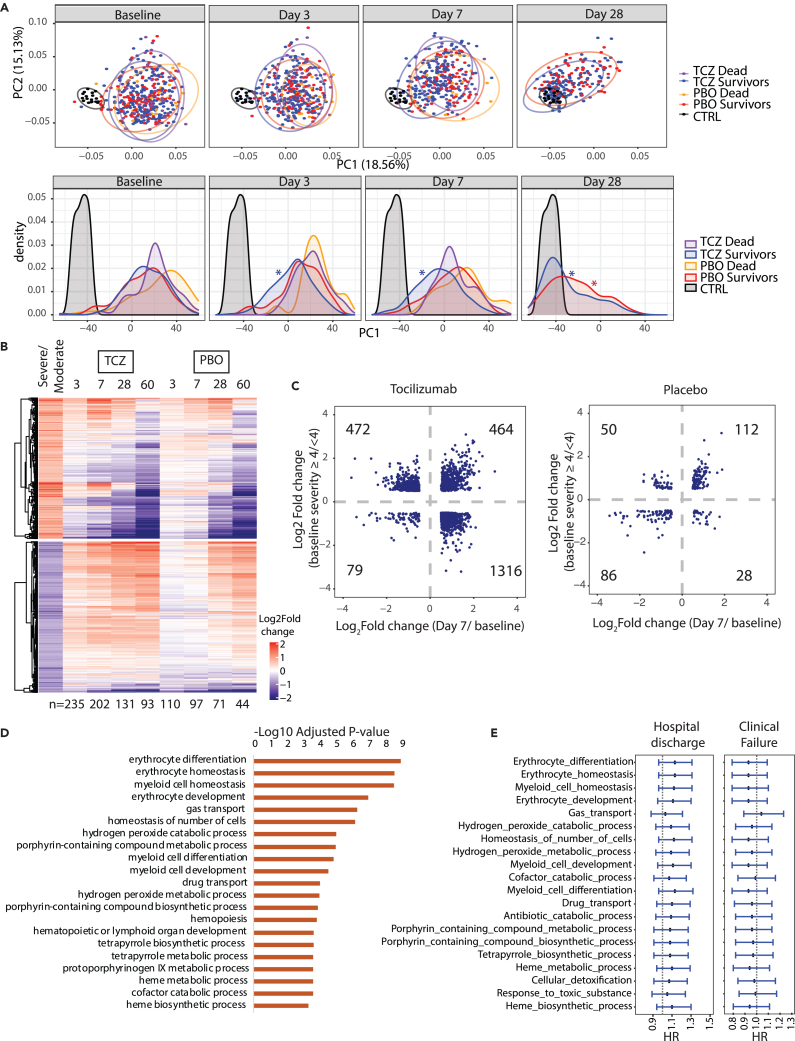
Figure 5.
Differential gene expression in response to tocilizumab treatment in hospitalized COVID-19 patients
(A) PCA and corresponding density plots showing longitudinal changes in RNA-seq signal at baseline, day 3, day 7, and day 28. ∗p value < 0.05, Kolmogorov-Smirnov test performed between the indicated timepoints relative to baseline.
(B) Heatmap showing longitudinal response of the blood transcriptome to TCZ treatment. Rows represent severity-associated genes identified by DESeq2. For each timepoint only samples measured at baseline and at the indicated timepoint were used for analysis (n indicated below the heatmap).
(C) Scatterplot of log2fold difference between severe vs. moderate baseline cases and day 7 after treatment vs. baseline for TCZ (left) and Placebo (right). Data points represent genes significantly different by ≥0.5 or ≥-0.5 log2fold for both comparisons.
(D) Bar plot showing immune pathways enriched among genes expressed higher in severe vs. moderate cases at baseline that also show greater increase on TCZ treatment vs. placebo by day 7 ([TCZ Day 7 – TCZ Day 1] – [PBO Day 7 – PBO Day 1]).
(E) Cox proportional hazard modeling for pathways shown in panel D. Forest plots generated as in (C). CTRL, control; HR, hazard ratio; PCA, principal component analysis; PBO, placebo; TCZ, tocilizumab.