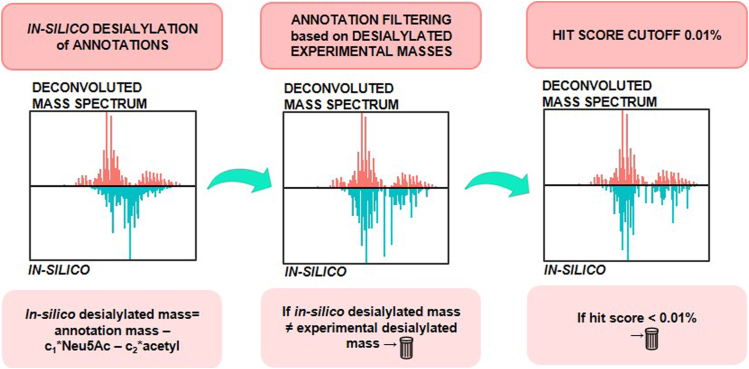
Fig. 5.
Schematic workflow of in silico filtering of intact glycoform annotations based on experimentally desialylated protein data. The upper panels show the deconvoluted experimental mass spectrum of r-hGAA after the neuraminidase digestion, while the lower panels show the simulated spectrum created upon in silico removal of sialic acids from all the potential MoFi annotations. Annotations were filtered based on a maximum mass error between the calculated and measured desialylated masses of 5 Da and subsequentially based on a hit score >0.01%. Magnified figures of the simulated spectra versus the experimentally desialylated spectrum are reported in supplemental Fig. S15. r-hGAA, recombinant human acid alpha-glucosidase.