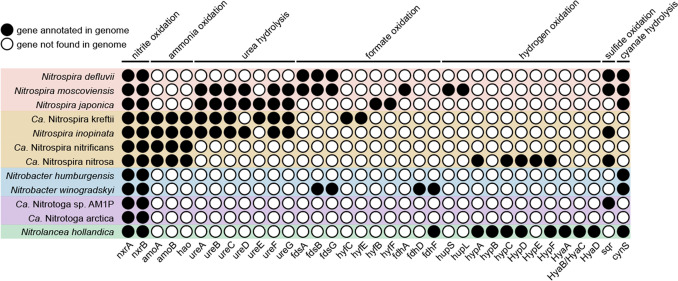
Figure 3.
Functional genes in NOB Nitrospira, comammox Nitrospira, Nitrobacter, Ca. Nitrotoga, and Nitrolancea based on mapped reads to the annotated ORFs. Twelve representative genomes that contained at least one of the related genes are displayed. Metagenemarks45 was used to predict the genes of each genome, and functional annotation referred to KEGG,46 COG,47 and Uniprot48 databases. Notably, due to differences in MAG quality and limitations of databases we used, it is possible that some genomes missed functional genes. As gene expression is regulated by various complex factors, such as environmental conditions, nutritional status, and intra- and extracellular signals, gene annotation only represents the potential for functional expression of these genes and cannot ensure that they will be expressed in the actual environment, which requires further investigation.