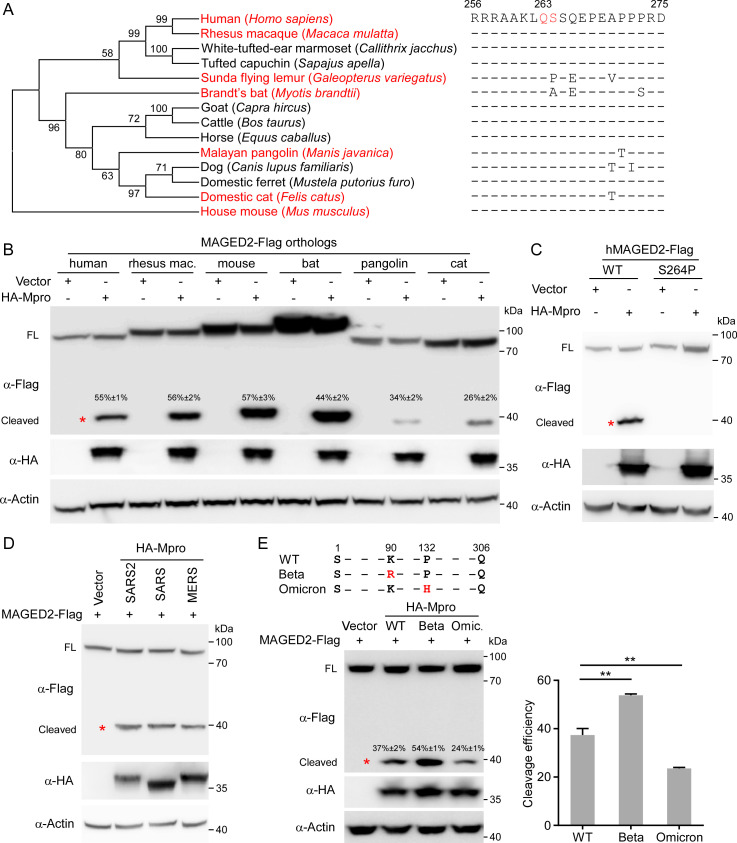
Fig 2.
MAGED2 cleavage by Mpro is conserved in multiple mammalian species and coronaviruses. (A) A phylogenetic tree was constructed based on the protein sequences of MAGED2 orthologs by using the neighbor-joining method conducted in program MEGA6. MAGED2 residues neighboring Mpro cleavage site from human, rhesus macaque, White-tufted-ear marmoset, tufted capuchin, Sunda flying lemur, Brandt’s bat, goat, cattle, horse, Malayan pangolin, dog, domestic ferret, domestic cat, and house mouse are aligned. (B to E) HEK293T cells were transfected with MAGED2 orthologs as indicated species. The uncleaved or cleaved protein band intensity was quantitatively analyzed using ImageJ. Cleavage efficiency = cleaved products/(cleaved products + uncleaved protein) × 100% (B), MAGED2 mutant S264P (C), and HA-tagged Mpro from SARS-CoV-2, other coronavirus (SARS-CoV or MERS-CoV) (D), or SARS-CoV-2 variants (Beta or Omicron) (E). Lysates of transfected cells were analyzed by immunoblotting with the antibodies indicated on the left. Western blots are quantified with ImageJ. Each experiment was independently repeated three times with similar results, and the representative images are shown. Values are means plus standard deviations (error bars) from one representative experiment with three biological replicate samples. **, P < 0.01 by one-way analysis of variance.