ABSTRACT
High error rates of viral RNA-dependent RNA polymerases lead to diverse intra-host viral populations during infection. Errors made during replication that are not strongly deleterious to the virus can lead to the generation of minority variants. However, accurate detection of minority variants in viral sequence data is complicated by errors introduced during sample preparation and data analysis. We used synthetic RNA controls and simulated data to test seven variant-calling tools across a range of allele frequencies and simulated coverages. We show that choice of variant caller and use of replicate sequencing have the most significant impact on single-nucleotide variant (SNV) discovery and demonstrate how both allele frequency and coverage thresholds impact both false discovery and false-negative rates. When replicates are not available, using a combination of multiple callers with more stringent cutoffs is recommended. We use these parameters to find minority variants in sequencing data from SARS-CoV-2 clinical specimens and provide guidance for studies of intra-host viral diversity using either single replicate data or data from technical replicates. Our study provides a framework for rigorous assessment of technical factors that impact SNV identification in viral samples and establishes heuristics that will inform and improve future studies of intra-host variation, viral diversity, and viral evolution.
IMPORTANCE
When viruses replicate inside a host cell, the virus replication machinery makes mistakes. Over time, these mistakes create mutations that result in a diverse population of viruses inside the host. Mutations that are neither lethal to the virus nor strongly beneficial can lead to minority variants that are minor members of the virus population. However, preparing samples for sequencing can also introduce errors that resemble minority variants, resulting in the inclusion of false-positive data if not filtered correctly. In this study, we aimed to determine the best methods for identification and quantification of these minority variants by testing the performance of seven commonly used variant-calling tools. We used simulated and synthetic data to test their performance against a true set of variants and then used these studies to inform variant identification in data from SARS-CoV-2 clinical specimens. Together, analyses of our data provide extensive guidance for future studies of viral diversity and evolution.
KEYWORDS: SARS-CoV-2, influenza, genomics, bioinformatics
INTRODUCTION
Large population sizes, high replication rates, and error-prone polymerases all contribute to the generation of sequence diversity found in viral infections (1 - 5). Natural selection acts on this diversity, contributing to viral evolution. RNA viruses have some of the highest mutation rates among viruses (1, 6, 7). To replicate their genomes, RNA viruses must encode their own RNA-dependent RNA polymerases, which often lack proofreading capabilities. Coronaviruses are a notable exception, as they possess a distinct protein with 3′–5′ exonuclease capability (1, 8, 9). Most errors made during replication—up to 40% in RNA viruses—are lethal (10, 11). Beneficial mutations make up a much smaller proportion, and these, along with neutral mutations, comprise the substitution rate. This substitution rate can be used to estimate the viral evolutionary rate, an important calculation in considering viral spread, pandemic potential, and vaccine design (4, 12).
Due to the large population sizes of RNA viruses, intra-host bottlenecks, and genetic drift, genetic diversity within a host is dynamic, with frequencies of mutations constantly rising and falling (13). Mutations can lead to changes in the consensus sequence, e.g., where the allele frequency (AF) is greater than 50%, and these specific sets of mutations separate globally circulating virus populations into clades. Mutations in the virus genomes that are not the majority within an infected host (i.e., present at lower than 50% frequency) represent minority variants. Deep sequencing enables the capture of intra-host variation, both at the majority and minority level, enabling the identification of variants and estimation of their frequency. Studying intra-host variation can help in tracking viral spread, estimating population bottleneck sizes, and identifying key amino acid changes that differentiate new viral strains (14 - 17). Additionally, minority variants can highlight regions of the genome under selection or regions with increased mutational tolerance, as well as allow for detection of subtle population shifts within the infected host and discovery of possible drug resistance mutations (18, 19). Thus, information gleaned from studying intra-host viral diversity has major implications for vaccine, monoclonal antibody, and drug development.
Given the many applications of studying intra-host viral diversity, accurately identifying and quantifying viral variants is essential. Precise identification of viral variants, especially those at low frequencies, is complicated by the fact that viral genome sequencing often requires reverse transcription and amplification, which, along with library preparation and the sequencing process, are error prone. Thus, distinguishing true sequence variation from technical and experimental noise is challenging. Typically, several ad hoc metrics are used to filter variants, such as applying frequency and coverage cutoffs to sequencing data; however, the frequency at which identified variants are considered valid can vary widely (20 - 26). Most studies using large sample cohorts, or performing analyses on publicly available data, generally use single replicate data, despite evidence suggesting that replicate sequencing may be essential for filtering false-positive minority variants (21). Despite the large number of studies analyzing minority variants in virus data, there is no consensus on what coverage cutoffs and AF cutoffs to use, and no large-scale studies have been performed to determine what thresholds lead to the highest confidence in variant identification.
In addition to the diversity of cutoffs used for single-nucleotide variant (SNV) identification, there is also great diversity in the variant-calling software available. Variant callers are often designed with specific functions in mind, such as identifying germline or somatic mutations in cancer genomes or SNVs in viral populations (27, 28). The function for which a variant caller is designed can have a significant impact on the statistics used and assumptions made by the software. Tools designed for detection of germline mutations, such as HaplotypeCaller (hc) and freebayes, must consider the very large reference genome, higher frequency variants, the diploid nature of the genome, the possibility of copy number variation, and long repetitive regions or large insertions or deletions (29 - 34). In these instances, local realignment of haplotypes may be most effective (27). By contrast, software used for somatic mutations in tumors, such as Mutect2 and Varscan, or for viral diversity, such as iVar and timo (a variant caller developed in our lab), use base-by-base comparisons, or a combination of this with haplotype-based alignment, to find lower frequency variants (21, 30, 32 - 35). These tools also may need alternative methods to reduce false-positive calls to account for PCR errors introduced during amplification of the viral genome (28). Due to the differences in bioinformatic and statistical approaches used by each variant-calling tool, identifying the tool that is the best fit for the specific research question being studied is essential. Some tools have been tested in pairwise comparisons (21, 34); however, little work has been done to extensively test the performance of many available tools on different viruses, across sequence coverages, and at various allele frequencies in viral deep sequencing data.
Here, we tested seven variant callers on simulated, synthetic, and clinical deep sequencing data. We tested each tool across a range of coverages, allele frequencies, and experimental designs to determine the optimal parameters that should be used to decrease false-positive variant identification, without sacrificing true-positive data. To compare performance between a small RNA virus with a high mutation rate, and a large RNA virus with proofreading capability, we tested the variant callers on two viruses of particular interest in the viral diversity field, influenza virus and SARS-CoV-2. We find that choice of variant caller and use of replicate sequencing have the most significant impact on SNV discovery and demonstrate how both allele frequency and coverage thresholds impact both false discovery rate (FDR) and false-negative rate (FNR). We also provide guidance on best practices for leveraging deep sequencing data from public repositories for intra-host studies. These analyses provide a resource for studies aiming to assess intra-host viral diversity in SARS-CoV-2 or influenza virus, and lay the groundwork for similar studies in other viruses.
MATERIALS AND METHODS
Extended methods are available in the supplementary materials.
Generation of simulated data
Reads were simulated using NEAT (v2.0) by constructing a mutation, error, fragment length, and guanine-cytosine (GC) model for each viral type (36). The models were provided to NEAT gen_reads.py along with reference fasta files and a mutation rate of 0.009 (0.9%) for influenza virus and 0.0045 (0.45%) for SARS-CoV-2 to produce a “golden” variant call format (VCF) file containing a defined number of SNVs in each virus. Simulated random PCR errors were also added to each replicate using gen_reads.py (NEAT). Several copies of the replicate golden VCFs were made, each with the same variants but with differing allele frequencies. These VCFs were used to simulate paired end fastq libraries at 100,000× genome coverage, and downsampling was used to simulate lower coverages.
Sequences were trimmed using trimmomatic v0.36 (37), aligned to the respective reference genome with BWA mem v0.7.17 (38), and duplicate reads were marked using GATK MarkDuplicatesSpark v4.1.7.0 (39). Variants were called in each replicate with seven different tools, using multiple parameter configurations for each tool (Table S1). A VCF file containing the intersection of the two replicates was generated using bcftools isec (v1.9) (38). The nextflow pipeline used for data simulation, sequence processing, variant calling, and analysis is available at https://github.com/gencorefacility/MAD2.
Synthetic RNA generation, library preparation, and data processing
Synthetic influenza A/H1N1pdm, “wild type” (WT), and variant RNA (created by adding 18, 14, and 14 nucleotide changes into the WT PB2, HA, and NA segments, respectively) were synthesized as double-stranded DNA (gBlocks) (sequences and details in Supplemental Methods). In vitro transcription with the HiScribe T7 High Yield RNA Synthesis Kit (Invitrogen) was used to generate full-length synthetic negative-sense gRNA. RNA samples were diluted to approximately equal concentrations (~6 × 108 copies/µL). The three segments of WT and variant RNA were mixed at approximately equal molarity to generate a master-mix. This master-mix was then mixed at different proportions of variant to WT RNA (1:2, 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:256) and serially diluted to 6 × 103 copies/µL. Samples from 103 to 106 were sequenced and used for analyses. Comparison made across copy numbers was done from 106 to 104, which had the most complete data sets and is specified in the figure legends.
cDNA was generated, and libraries were prepared using the Nextera XT library preparation kit (Nextera), with all volumes scaled down to 0.25× of the manufacturer’s instructions, cleaned with AMPure beads, and pooled at equal molarity. Libraries were sequenced on the Miseq 300 Cycle v2 using 2 × 75 pair-end reads. Samples were amplified and sequenced in duplicate and analyzed with the pipeline described above, with the addition of adapter trimming.
SARS-CoV-2 clinical sample preparation, processing, and variant calling
Total RNA was extracted from 300 µL of nasopharyngeal or mid-turbinate swabs collected at the National Institutes of Health (NIH) Clinical Center as part of diagnostic testing between 24 July 2020 and 31 March 2021 (Table S2). All samples were de-identified and anonymized.
RNA from samples was extracted using the NucliSENS easyMAG automated nucleic acid extractor, and the viral genome was amplified using a modified version of the ARTIC protocol (https://artic.network/ncov-2019), and the methods are described at https://github.com/GhedinSGS/SARS-CoV-2_analysis. All libraries were prepared as above and sequenced on either the Illumina MiSeq or the Illumnia NextSeq500 using either the 2 × 150 bp or 2 × 300 bp paired end protocol. All samples were processed in duplicate.
Samples were processed with the pipeline available and described above, with the addition of merging the two SAM files (from A and B primer pools) for each biological sample into one alignment file using Picard Tools MergeSamFiles v2.17.11. Variants were called as above using the standard parameters for each tool (Table S1). To confirm our findings, replicate SARS-CoV-2 sequencing data used in a within-host diversity study were downloaded (PRJEB37886, PRJEB42623) (40) and aligned to the Wuhan-Hu-1 reference genome (NC_045512.2) using Minimap2 (41). Minority variants were then called using iVar and timo with custom input parameters (Table S1).
RESULTS
Simulated and synthetic data provide a “true” set of minority variants to assess variant caller performance
To test the ability of each variant caller to accurately identify variants, it is essential to know the “true set” of variants within the data. With this in mind, we tested the ability of six popular variant-calling software packages (Freebayes, hc, iVar, Lofreq, Mutect2, and Varscan) and one in-house pipeline (timo) to accurately identify minority variants in simulated and synthetic sequencing data (Fig. S1) (21, 32 - 35). Single-nucleotide variants were simulated across three influenza virus genomes (A/H1N1, A/H3N2, and B/Victoria) and one coronavirus genome (SARS-CoV-2) at both defined and random allele frequencies and across a range of downsampled coverages (Fig. S1A and B). Furthermore, synthetic RNA controls of three influenza virus segments (PB2, HA, and NA) containing known SNVs (“variant”) were mixed with “wild-type” segments in varying amounts at various dilutions to create a range of allele frequencies and genome copy numbers (see Materials and Methods) (Fig. S1C and D). Combined, we used these synthetic and simulated data sets to test variant caller performance on a known set of SNVs.
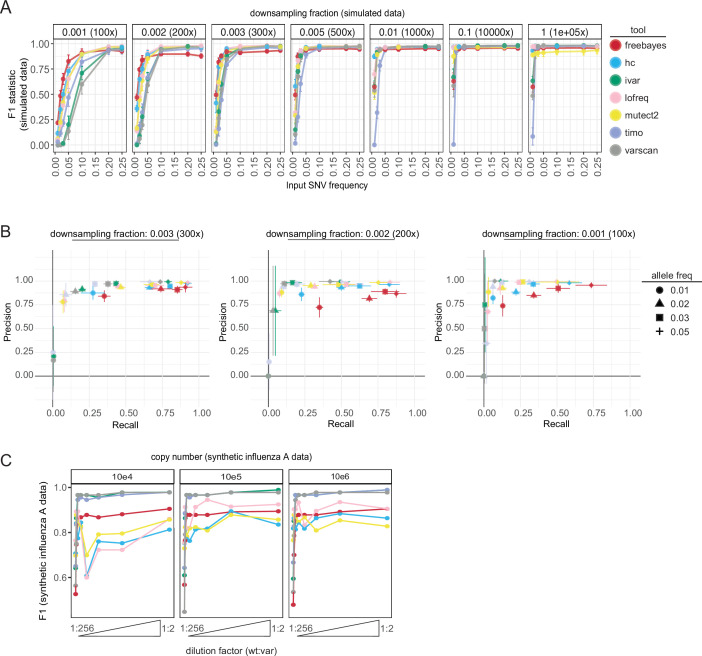
We found that all callers performed poorly on the simulated data using their default parameters (Fig. S2). Therefore, to compare all callers equally, we used a standard set of permissive input parameters [min coverage = 1×, allele frequency cutoff = 0.01 (1%)] throughout our testing (Table S1). When assessing the F1 statistic across a range of simulated frequencies, most variant callers performed better at low frequencies [≤0.05 (5%)] when the coverage was high (downsampling fraction ≥0.005 or ~500× expected read depth). Conversely, high frequencies were necessary for accurate variant detection at small downsampling fractions where the average coverage was low (Fig. 1A). A noticeable drop in performance was observed across most callers, particularly timo, at allele fractions of 0.01 (1%), even at the highest assessed coverage. A closer look at precision and recall for each tool at downsampling fractions 0.001 (~100× read depth), 0.002 (~200×), and 0.003 (~300×) indicated that tools trade recall for precision at frequencies between 0.01 and 0.05 (1%–5%) (Fig. 1B). Varscan, iVar, and timo tended to be extremely conservative, especially at allele frequencies of 0.01 (1%). Decreasing the input frequency parameter from 0.01 (1%) to 0.001 (0.1%) decreased the stringency of timo, allowing for more input SNVs to be identified, while the performance of Varscan and iVar was not impacted (Fig. S2 custom input parameters, Table S1).
Fig 1.
Variant caller performance on simulated and synthetic data. (A) F1 statistic for each variant caller on simulated data across a range of downsampling fractions (y-axis: 0.001–1, expected read depth: 100×–100,000×) and allele frequency values (x-axis: 0.01–0.25 or 1%–25%). Values shown are mean and standard deviation of the four viruses (A/H1N1, A/H3N2, B/Victoria, and SARS-CoV-2) using standard input parameters (Table S1). Color represents the variant caller used. (B) Precision (y-axis) and recall (x-axis) graphs of each variant caller across allele frequencies 1%–5% (point shape) for downsampling fractions 0.003 (~300× read depth, left), 0.002 (~200×, middle), and 0.001 (~100×, right). Color represents variant caller. Mean and standard deviation are shown across the four viruses for precision and recall scores. (C) F1 statistic (y-axis) for each variant caller using standard inputs on synthetic influenza A virus data across a range of copy numbers (104–106) and dilutions (wt:var—1:256, 1:128, 1:64, 1:32, 1:16, 1:8, 1:4, 1:2). Data are grouped across the PB2, HA, and NA segments to calculate F1. Color represents the variant caller used.
Simulated data lack the reverse transcription, amplification, and sequence library preparation steps involved in the generation of data from clinical specimens. To assess how these sample preparation steps, along with duplicate sequencing, and SNV thresholds may impact variant caller performance, we tested each tool on the synthetic influenza A virus (IAV) RNA data set (Fig. 1C; Fig. S1 C and D). The mean and median read depth across gene segments were greater than 1,000× and had similar coverage distributions to our simulated data sets at downsampling fractions of 0.01 (~1,000×) and 0.1 (~10,000×) (Fig. S1E). As observed in the simulated data, F1 statistics were highest when the variant (var) gene segments were present at higher proportions (dilutions ≥1:32) within the sample (Fig. 1C). Freebayes, Lofreq, HaplotypeCaller, and Mutect2 were the most influenced by copy number and had a notable drop in performance when used on the synthetic IAV data compared with the simulated data sets—demonstrating the importance of testing variant caller performance across multiple data types.
Frequency thresholds and sequencing replicates reduce false-positive SNVs
Previous studies have reported the necessity of establishing frequency and coverage thresholds as well as having replicate sequencing to decrease false-positive SNVs in a data set (27, 40). Given that most publicly available data consist of single replicate sequencing data, we aimed to establish coverage and frequency thresholds that would minimize the FDR and FNR to levels comparable with those observed using two replicates. To do this, we used both simulated and synthetic data sets with standard input parameters and ignored the “binocheck” requirement from timo (which requires variants to be found in both forward and reverse reads consistent with binomial sampling), allowing us to test the performance of timo on low frequency SNVs.
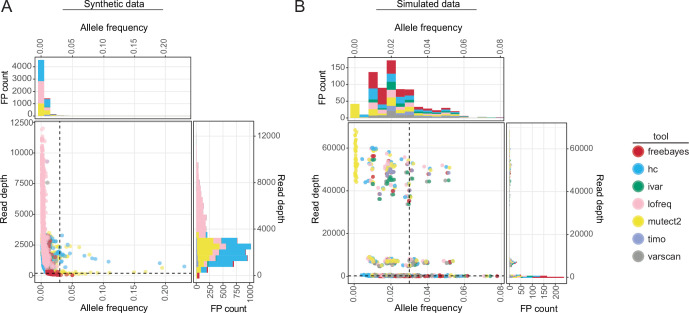
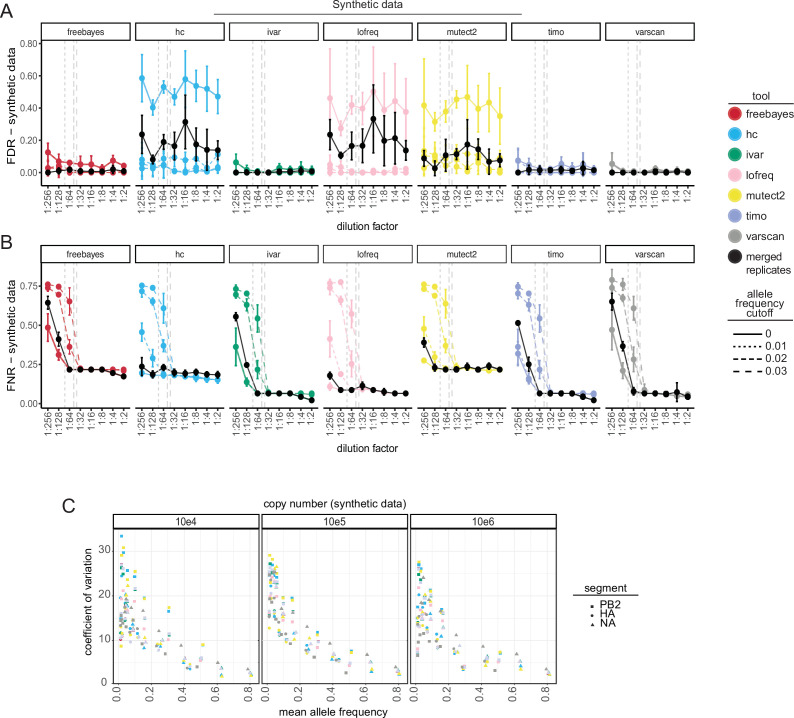
False-positive SNVs were found across a range of output read depths in both the synthetic (Fig. 2A, 40×–11,995×) and simulated (Fig. 2B; Fig. S3A, 1×–68,441×) data. Therefore, applying coverage cutoffs of 100×–300× did not drastically impact the number of false-positive calls in either the simulated or synthetic data sets (Fig. S3). However, coverage is important when considering SNV recall (Fig. 1B). Given that false-positive SNVs were primarily identified at allele frequencies less than 0.03 (3%) (Fig. 2A and B), applying frequency thresholds to single replicate data lowered the false discovery rate for all callers (Fig. 3A; Fig. S4 A). However, using frequency thresholds did come at the cost of significantly increasing the FNR, especially when using 2% and 3% cutoffs, as true SNVs found at low frequencies were filtered from the data (Fig. 3B; Fig. S4 B). In contrast, keeping only SNVs shared between the two replicates dramatically decreased the FDR while maintaining relatively low FNRs (Fig. 3A and B; Fig. S4 A and B). The majority of false-positive SNVs that remained in the synthetic data after merging replicates was present at low frequencies (dilution factors 1:256, 1:128). Therefore, using an allele frequency cutoff of 1% (0.01) on replicate sequencing data can further increase the confidence in SNV calls. Replicates also increased the accuracy of allele frequency estimation of true-positive variants found in the simulated data, especially for SNVs in low coverage data, where the percent error of allele frequency estimation is pointedly lower for all tools when using replicates (Fig. S5A). HaplotypeCaller, Lofreq, and Mutect2 called notably higher numbers of false-positive SNVs in synthetic data, including many that were maintained even after merging replicates—indicating that these callers make consistently incorrect SNV calls (Fig. 1C; Fig. 2A; Fig. 3A). Furthermore, these three callers had multiple instances where true-positive SNVs were identified at high frequencies (AF >0.05) in one replicate and were entirely absent in the other (Fig. S5B).
Fig 2.
The output frequency and coverage of false-positive variants in synthetic and simulated data. (A and B) Scatter plots and associated histograms showing the number of false-positive SNVs identified at different output allele frequencies and total read depths for all callers and copy numbers (103–106) in the synthetic influenza A virus samples (A) or across all callers, viruses, and downsampling fractions in simulated data (B). Dotted lines are drawn at allele frequency = 0.03 and read depth = 200×. Color represents the variant caller used.
Fig 3.
Effect of frequency cutoffs and sequencing replicates on variant detection and quantification in synthetic influenza A virus data. (A) False discovery rate (FDR) ( ) and (B) false negative rate (FNR) ( ) of synthetic influenza A virus data as a function of dilution factor using either single replicate data (colored points and lines) with applied frequency cutoffs (line type) or merged two replicate data without cutoffs (solid black points and lines). Variants below the applied frequency cutoff are filtered out and considered false negatives. Dashed vertical lines indicate the location of allele frequency cutoffs relative to the dilution factors. Values shown are the mean and standard deviation across all sequenced copy numbers (103–106). FP: false positive, TP: true positive, FN: false negative. (C) Coefficient of variation (y-axis, ) of synthetic influenza A virus data across a range of copy numbers (104–106) vs the mean allele frequency (x-axis) within a segment and dilution factor. Only true-positive variants found across all variant callers were considered in this analysis. Color represents the variant caller used. Point shape indicates the synthetic gene segment.
The synthetic IAV data are especially well suited for testing the amount of variability associated with allele frequency estimation due to various experimental factors. As a property of the design, all variants on a segment are linked, and thus, the true allele frequencies are expected to be identical. By measuring the amount of variation in allele frequency of the variants across each segment, we can determine which factors influence this estimation the most. We find that copy number does not affect the variation in allele frequency estimation (Fig. 3C). By contrast, the frequency of the variant has a pronounced effect on the accuracy of allele frequency estimation. The lower the allele frequency, the higher the variance in the estimation as determined by the coefficient of variation further justifying the use of an allele frequency cutoff of 1% (0.01) in variant analyses. Notably, variants located at the end of the gene segments (PB2 pos: 2280 and HA pos: 4), where coverage was low (Fig. S1E), or variants next to each other (PB2 pos: 2266, 2267, HA pos: 515, 516, and NA pos: 282, 283, 284) were frequently missed by the tools or were found at aberrantly low frequencies when detected (Fig. S1E and S5C).
Together, these results highlight the factors that influence the accuracy of identifying and quantifying variants and indicate that using replicate sequencing with less stringent frequency cutoffs (AF ≥0.01, 1%) is the best combination to reduce the FDR while maintaining a low FNR (Fig. 3A and B) and for accurate allele frequency estimations (Fig. 3C; Fig. S5 A). However, when replicate sequencing is unavailable, strict read depth (≥200×) and frequency (AF ≥0.03, 3%) cutoffs are necessary (Fig. 3A and B; Fig. S3A through D; Fig. S4A and B).
Choice of variant caller significantly impacts set and frequency of identified variants in real SARS-CoV-2 data using single replicate data
While simulated and synthetic data allow for testing minority variant callers and cutoffs in a controlled setting, real data will always be more unpredictable. Thus, after using simulated and synthetic data to assess variant caller performance across frequencies and coverages, we tested how the callers performed on SARS-CoV-2 sequence data from diagnostic samples. Based on the simulated and synthetic data testing, we determined that a coverage cutoff of 200× and an allele frequency cutoff of 0.03 (3%) in single replicate data minimized false-positive calls without sacrificing large amounts of true-positive data with most variant-calling tools (Fig. 3A and B; Fig. S3A through D; Fig. S4 A). To test the variant-calling tools on high-quality data, we used only samples where at least 80% of the genome had a read depth over 200× coverage cutoff in both sequencing replicates (Fig. S6A). We used each variant-calling tool to identify minority variants in these samples and filtered them using a read depth cutoff of 200× and an allele frequency cutoff of 0.03 (3%).
We were interested in how similar the sets of identified variants were across each caller. As a proof of principle, we filtered the set of variants for those present above an allele frequency of 0.5 and at read depths greater than 5× to identify consensus changes (AF ≥ 50% or major variants) within the data. As expected, the tools largely agreed on the consensus changes within the data (Fig. S6B). There was a small set of major variants that the callers did disagree on; however, most of which were a result of differences in the way some callers identify indels or handle variant at consecutive nucleotide positions. For the purposes of this study, indels were excluded from the analysis. These data indicate that even at high allele frequencies, the variant callers disagree to some extent on the set of variants present in clinical data, an important consideration when choosing how to define consensus sequences from SARS-CoV-2 data.
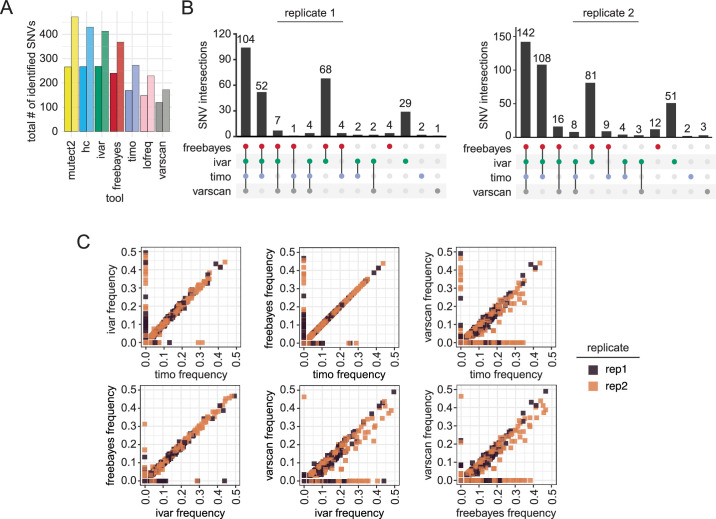
We then analyzed the intersection of the minority variants (AF between 3% and 49%) identified by each tool. The total number of variants identified varied greatly between the callers, with Varscan calling the fewest variants, followed by timo and Lofreq, in line with the more conservative nature of these callers observed in the previous analyses (Fig. 4A). Of note, we found that replicate 2 data had much higher numbers of minority variants, particularly at very low frequencies, regardless of the cycle threshold (Ct) value or date of sequencing. This suggests that freeze thawing samples may impact minor variant numbers (Fig. S6C) (42, 43). When comparing the set of minority variants identified by each of the seven tools, there was significant disagreement between the variants. Mutect2 and HaplotypeCaller identified many variants that other callers did not, particularly in replicate 1, and missed several variants identified by the other callers (Fig. S6D). This was similar to the performance of these callers on the synthetic data sets. Given the high number of false positives identified by HaplotypeCaller, Mutect2, and Lofreq in the simulated and synthetic data sets, we focused on the intersection of minority SNVs found in just the other four variant callers: Freebayes, iVar, Varscan, and timo. Of all the minority variants found in the data, 104 from replicate 1 and 142 from replicate 2 were identified by all four of the variant callers (Fig. 4B). Overall, choice of variant caller appears to have a significant impact on the set of minority variants identified in SARS-CoV-2 data from clinical specimens.
Fig 4.
Effect of variant caller on identification and allele frequency estimation of SNVs in SARS-CoV-2 data from clinical samples. (A) Bar plot showing raw number of minor variants identified by each variant caller in replicate 1 (left bar) or replicate 2 (right bar) using a 3% allele frequency cutoff. (B) UpSetR plot showing agreement of minority variants in each replicate across Freebayes, iVar, timo, and Varscan using an allele frequency cutoff of 0.03 (3%) and coverage cutoff of 200×. Vertical bars indicate the size of the shared set of variants, while dots and connecting lines show which callers share a given set of identified variants. (C) Scatter plot showing the output frequency of minority variants identified by two different variant callers. Color represents replicate. Variants with frequency of 0 were not identified by that variant caller.
Many studies of minority variants investigate the frequency of minority variants to calculate selection, bottleneck size, and potential for transmission (22, 44). We were interested in how well the variant callers agreed on the frequency at which variants were identified. We plotted the frequency of a variant in one caller against the frequency in each other caller and found that most of the minority variant callers were strikingly similar in their frequency calls of shared variants. Timo, Freebayes, and iVar all showed almost complete agreement on the frequency of shared variants (Fig. 4C), with Freebayes and iVar having the best agreement of SNVs found ≥20%. Varscan showed more variation in frequency, generally calling variants at a lower frequency than the other three tools (Fig. 4C). Of interest, variants called by one caller but not another spanned a frequency range of 0.03 (3%) all the way to 0.5 (50%), indicating that even high-frequency minority variants were often not agreed upon by variant callers. These data show that choice of variant caller not only affects the set of the minority variants that are identified in a data set, but also the frequency of those variants.
Most minority variants in data from SARS-CoV-2 clinical specimens are not reproducible across sequencing replicates
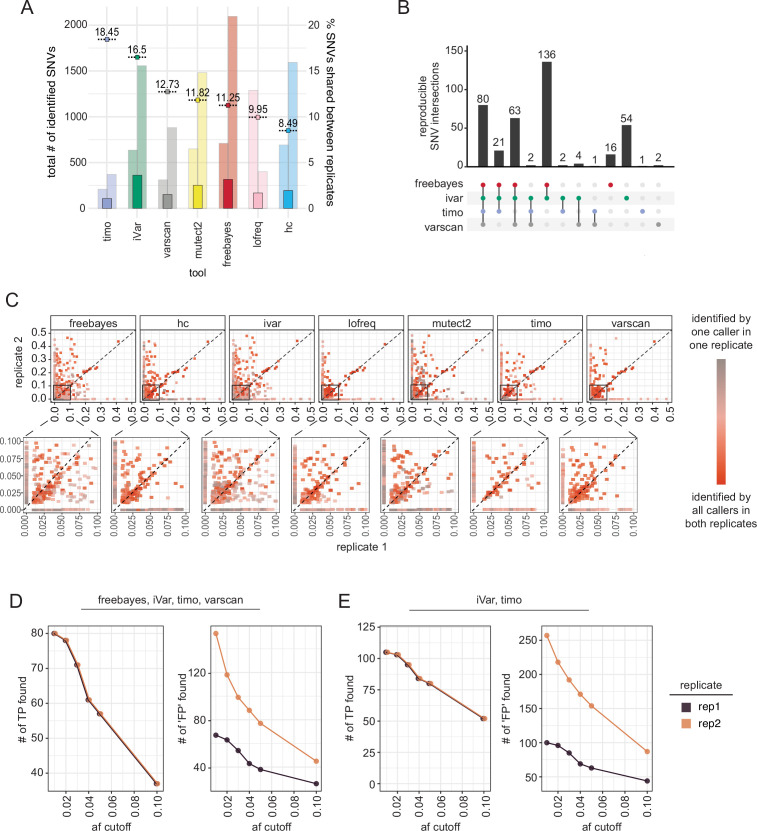
In our sequencing data, the number of variants identified in each replicate by each tool was markedly different, suggesting that many of the identified minor variants may not be true variants introduced through viral replication but instead technical artifacts (Fig. 4A; Fig. S6 C and D). As was shown with our simulated and synthetic data, errors introduced through PCR, library preparation, and sequencing are mostly random and therefore less likely to reappear and be identified across multiple sequencing replicates, particularly when using Freebayes, iVar, timo, or Varscan (Fig. 3A). To find high-confidence minority variants, we looked at the intersection of variants between the two replicates using each caller and a 0.01 (1%) allele frequency threshold, as established in synthetic data for merged replicates (Fig. 3A and B). iVar and Freebayes called the highest number of reproducible variants, while timo called the fewest number of reproducible variants (Fig. 5A). However, out of the total variants identified between the two sequencing replicates, timo had the highest percentage of reproducible variants (18.45%) suggesting that being conservative may lead to increased reproducibility between replicates and an increased confidence in SNV calls when used on single replicate data. It is, however, important to note that the relatively low percentages of reproducible variants are likely skewed by the high numbers of low-frequency variants found in replicate 2 (Fig. 5A; Fig. S6 C). When we looked at the intersection of only the variants found by the tools in both replicates, less than a third were found by the callers across replicates, suggesting again that variant callers do not agree on the set of minority variants present (Fig. 5B). Together, these data suggest that most minority variants are not reproducible across replicates and support the idea that more than any other criteria, sequencing replicate has the highest impact on the set of minority variants identified (Fig. 5B; Fig. S6 C and D).
Fig 5.
Reproducibility of minority variants across sequencing replicates. (A) Bar plot showing number of reproducible minor variants across sequencing replicates by each variant caller using a 1% allele frequency cutoff. Percentages shown are the percentage of total individual variants that were reproducible. Background bars indicate the total number of variants found by each tool in each replicate (left: replicate 1, right: replicate 2). Data are sorted by percentage of shared SNVs. (B) UpSetR plot showing overlap of reproducible variants across Freebayes, iVar, timo, and Varscan, using a frequency cutoff of 0.01 (1%) and coverage cutoff of 200×. Vertical bars indicate the size of the shared set of variants, while dots and connecting lines show which callers share a given set of reproducible variants. (C) Scatter plot showing frequency of variants across sequencing replicates with frequency in replicate 1 on the x-axis and frequency in replicate 2 on the y-axis. Color represents reproducibility of each variant across variant callers and replicates. Inset highlights variants found at allele frequencies ≤0.10 (10%) in both replicates. The dotted line represents the x = y-axis and indicates perfect agreement between replicates. (D, E) Line graph showing the number of “true-positive” and “false-positive” variants in single replicate data across allele frequency cutoffs for all tools (D) or just iVar and timo (E). A true positive (TP) variant is defined as an SNV found by the selected callers in both replicates [80 variants shown in (B)], and a false positive (FP) is defined as any other variant found in an individual replicate by the selected callers. Color represents sequencing replicate.
Variants identified by all variant callers show the most reproducible frequencies
Using synthetic data, we showed that in a controlled setting, SNVs that were found in both sequencing replicates generally showed reproducible frequencies (Fig. S5B). Given that the frequency is an important metric in most analyses performed using minority variant data, we wanted to test if this held true in clinical samples. While some variants showed consistent frequencies, others differed drastically—identified at 5%–10% in one replicate and as high as 45%–50% in the other replicate (Fig. 5C). These data were striking as they reveal that averaging frequency across replicates, or performing only one sequencing replicate, could drastically alter downstream analyses performed using these numbers. Interestingly, when we looked at the variants that were reproducible across replicates and found by most, or all the variant callers, frequency tended to be much more consistent than those identified only in a single replicate, or by a single caller (Fig. 5C, dark red points). Together, these data suggest that confidence in each variant and its frequency is increased with replicate sequencing and identification by many variant callers.
Since replicate sequencing data are not always available, we investigated what frequency cutoff could be applied such that single replicate data closely resembled the merged replicate data. To do this, we looked at the intersection of SNVs called in both replicates by Freebayes, iVar, timo, and Varscan (80 variants out of 382 shown in Fig. 5B) and compared those with the intersection of SNVs called by the same four callers in each individual replicate (Fig. 4A). We then applied allele frequency cutoffs between 0.01 and 0.1 (1%–10%) to determine the best cutoff for use on single replicate data. Here, we identify a true positive as a variant present in the reproducible set and a false positive as any other variant found in a single replicate. As was noted previously, we find that replicate 2 data show an increased number of SNVs, perhaps due to freeze/thawing of samples between preparations (Fig. 5D; Fig. S6C). As such, replicate 1 is likely more representative of what single replicate data may typically look like. At an allele frequency cutoff of 0.01 (1%), all true positives were found, but the number of false positives was very high, while a frequency cutoff of 0.05 or 0.1 (5%–10%) removed an outsized number of true positives from the data set (Fig. 5D). Based on these data, we suggest an allele frequency cutoff of 0.03 (3%) when only single replicate data are available, a cutoff that was also confirmed in the simulated and synthetic data sets (Fig. 2A and B; Fig. 3A; Fig. S4A and B). We further suggest using the intersection of multiple variant callers to increase confidence in the data, especially when estimating SNV frequency (Fig. 5C). Using all variant callers for analysis would likely be tedious and unrealistic, thus we looked at the intersection of just two callers, iVar and timo, and we found a similar trade-off in true-positive and false-positive data when using a single replicate and a cutoff of 0.03 (3%) (Fig. 5E).
To determine if the discordance between replicate SARS-CoV-2 sequencing data is a common issue, we expanded our analyses to 1,181 SARS-CoV-2 samples that were sequenced in duplicate and used in a within-host diversity study (40) (Supplemental Methods). Samples used for SNV analyses were required to have at least 200× read depth across 80% of the genome in both sequencing replicates (Fig. S7A), which left 227 samples for minority variant analyses (Fig. S7A, inset). In addition, we limited our analyses to only timo and iVar outputs using custom input parameters (Table S1). Sequencing replicates shared anywhere from 0% to 40% of identified SNVs (Fig. S7B), with timo comparisons often having higher fractions of shared SNVs.
Using the Tonkin-Hill et al. (40) data set, we tested the impact of filtering using our suggested thresholds for single replicate data (output by both timo and iVar, ≥3%, ≥200×) and replicate data (in both replicates, ≥1%, 200×). The number of identified SNVs in the single replicate comparisons (replicate 1: 762 SNVs, replicate 2: 515 SNVs) was lower than when taking the intersection of both replicates (timo: 2,682, iVar: 3,108) (Fig. S7C). The decrease in total SNVs is likely due to our stringent 3% frequency requirement for single replicate data. However, approximately 84% (643/762) and 95% (487/515) of SNVs in replicates 1 and 2, respectively, were also found when taking the intersection of sequencing replicates (Fig. S7C).
Based on these data, it is clear that there are many considerations necessary when performing minority variant analyses, and parameters and cutoffs should thus be chosen carefully and thoughtfully, depending on the data available. In general, using replicate data and multiple callers provides the highest confidence set of SNVs and the most accurate frequency estimates.
DISCUSSION
It has long been understood that intra-host viral populations are heterogeneous in nature; however, capturing and measuring this viral diversity is complicated due to errors introduced during preparation and sequencing. We set out to identify the optimal tools, parameters, and filtering methods necessary for accurate variant identification. To accomplish this goal, we used a combination of simulated and synthetic sequence data to test the technical and experimental challenges and limitations of minority variant analyses. We found that sequencing depth and choice of variant caller have a significant impact on sensitivity of minor variant calls. Additionally, our results show that replicate sequencing allows for the use of lower frequency thresholds, and this combination provides the best results, keeping the false discovery rate low, without sacrificing true-positive data. Using replicates also decreases the error associated with estimating allele frequency in both simulated and synthetic data, although very low-frequency variants may still elude highly accurate estimates.
Using a standardized set of parameters, most callers performed relatively similarly on high coverage simulated data, having both high precision and high recall. The main differences in caller performance were seen in lower coverage data or at low frequencies. As many minority variants are found at low frequencies, understanding how tools perform under these conditions is more relevant to analyses of real sequencing data. Timo had the lowest recall at lower coverages and simulated frequencies due to its rigid requirements for SNVs to be above the 0.01 threshold parameter, while many other callers found SNVs at or below this frequency, regardless of setting a 0.01 AF cutoff. Timo, iVar, and Varscan all have the functionality to drop the input frequency parameter down to 0.001 (0.1%). Decreasing this parameter did not change the accuracy of iVar and Varscan but did increase the recall of timo. These data highlight the importance of optimizing bioinformatic tools to one’s own data.
As previously observed by our group and others, the best method for filtering out errors generated during sample processing is to sequence each sample twice and only keep the SNVs found in both replicates. Sequencing replicates removed nearly all false-positive calls in simulated data and significantly reduced the number of false-positive SNVs in the synthetic data sets. However, for the synthetic data sets, the number of false-positive SNVs was highly dependent on the variant caller used. HaplotypeCaller, Lofreq, and Mutect2 were all made and optimized for identifying variants in cancer cell data sets and had significantly higher false discovery rates than tools designed for viral use, particularly at low allele frequencies. Adjusting the filtering or input parameters on these callers may better optimize them for their use on viral data. For example, HaplotypeCaller suggests additional filtering of output data; however, when applied to this data set, SNV detection was significantly reduced. Without this additional filtering, most variants are identified but high numbers of false positives are included, suggesting additional optimization could improve performance.
The design of the synthetic IAV data also allowed us to test the effect of genomic position on variant detection and allele frequency. In influenza virus sequencing data, the ends of the genomic segments routinely have lower coverage than internal regions, due to poor end-capture during sequencing. By engineering variants near the ends of the segments in the synthetic data, we found that variants at these positions are often missed entirely by the tools (false negatives), and when they are detected, their allele frequencies are poorly estimated. This suggests that variants found at the ends of segments and more generally, in low-coverage regions of the genome, should be interpreted with caution. We also engineered variants that were immediately adjacent to each other. These variants were also often missed by the tools, despite having comparable coverage with other variants that were detected as true positives. This may be due to the variant callers preferentially assigning consecutive nucleotide changes as indels, rather than SNVs, excluding them from these analyses. When taken together, it is clear that genomic position does affect the performance of bioinformatic tools and that an understanding of the underlying biology and technical procedures should be used to inform viral variant calling.
We tested the optimized frequency and coverage cutoffs using SARS-CoV-2 sequence data from clinical infections. Most variant callers did not agree on the set of minor variants in the virus sequence data from clinical samples, and most minority variants were not reproducible across replicates. Ultimately, we determined that using the more stringent variant callers (timo, iVar, and Varscan), sequencing replicates, and moderate allele frequency (≥1%) and read depth (≥200×) cutoffs provide the highest confidence in the output SNV calls and allele frequency estimations. However, when replicate sequencing is unavailable, we suggest using a more stringent frequency cutoff (≥3%) on SNVs identified by multiple variant callers.
Combined, the simulated, synthetic, and clinical data sets show that there will always be a trade-off between inclusion of the maximum number of true variants and inclusion of false-positive data. Our study provides an extensive framework for studying minority variants in sequence data from clinical samples, outlining major considerations around choice of variant caller, application of frequency and coverage thresholds, and use of replicate sequencing. Furthermore, we have established a pipeline that can be used for further testing and optimization of parameters, or for other viruses. This work will inform and improve future studies of intra-host variation and estimates surrounding viral diversity and viral evolution.
ACKNOWLEDGMENTS
This work was supported in part by the Division of Intramural Research (DIR) (E.G.) of the NIAID/NIH, by NIAID/NIH R01 AI140766 (D.G., S.S.C.), NIGMS/NIH R01GM134066 and R01GM107466 (D.G.), NIH/NIAID 75N93019C00052 (D.G., S.S.C), R01 AI148574 (M.J.M.), and by Deutsche Forschungsgemeinschaft grant SFB 1310 (M.Ł.). M.Ł. is a Pew Biomedical Scholar and was partially supported by the NIAID Centers of Excellence for Influenza Research and Surveillance (contract HHSN272201400008C). K.E.E.J. was supported in part by the Public Health Service Institutional Research Training Award T32 AI007180. M.K. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T32GM132037. This work utilized the computational resources of the NIH High Performance Computing (HPC) Biowulf cluster (http://hpc.nih.gov) and the NYU IT High Performance Computing resources, services, and staff expertise.
Contributor Information
D. Gresham, Email: dgresham@nyu.edu.
E. Ghedin, Email: elodie.ghedin@nih.gov.
J. S. Malik Peiris, University of Hong Kong, Hong Kong, China .
DATA AVAILABILITY
Synthetic influenza data (Bioproject PRJNA865369) and SARS-CoV-2 data from clinical samples are available in NCBI GenBank and SRA (Bioproject PRJNA857712). Accession IDs can be found in Table S2. All downstream analysis files are available at https://github.com/GhedinSGS/Optimized-Quantification-of-Intrahost-Viral-Diversity. R functions for performing SNV analysis and generating plots were compiled into an R package, vivaldi (Viral Variant Location and Diversity), available at https://cran.r-project.org/web/packages/vivaldi/index.html.
ETHICS APPROVAL
All samples were anonymized and obtained with consent as part of SARS-CoV-2 diagnostic testing.
DIRECT CONTRIBUTION
This article is a direct contribution from Elodie Ghedin, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Morgane Rolland, U.S. Military HIV Research Program, HJF, and Johannes Goll, The Emmes Company LLC.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01046-23.
Experimental setup of simulated and synthetic data generation.
Variant caller performance using default and custom parameters on simulated data.
Effect of coverage cutoffs and sequencing replicates on false discovery rate and false negative rate in synthetic and simulated data.
Effect of frequency cutoffs and sequencing replicates on false discovery rate and false negative rate in simulated data.
Effect of sequencing replicates and genome location on the accuracy of allele frequency estimation.
Quantification of majority and minority variants identified in data from SARS-CoV-2 clinical specimens.
Reproducibility of minority variants across sequencing replicates from the Tonkin-Hill et al. 2021 sequencing data set.
List of variant callers and parameters; list of diagnostic samples.
Details on some of methods.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Arnold JJ, Cameron CE. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43:5126–5137. doi: 10.1021/bi035212y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanjuán R. 2012. From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog 8:e1002685. doi: 10.1371/journal.ppat.1002685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323 [DOI] [PubMed] [Google Scholar]
- 4. Peck KM, Lauring AS. 2018. Complexities of viral mutation rates. J Virol 92:e01031-17. doi: 10.1128/JVI.01031-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Domingo E. 2002. Quasispecies theory in virology. J Virol 76:463–465. doi: 10.1128/JVI.76.1.463-465.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lynch M, Ackerman MS, Gout JF, Long H, Sung W, Thomas WK, Foster PL. 2016. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17:704–714. doi: 10.1038/nrg.2016.104 [DOI] [PubMed] [Google Scholar]
- 8. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith EC, Sexton NR, Denison MR. 2014. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol 1:111–132. doi: 10.1146/annurev-virology-031413-085507 [DOI] [PubMed] [Google Scholar]
- 10. Sanjuán R, Moya A, Elena SF. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A 101:8396–8401. doi: 10.1073/pnas.0400146101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Visher E, Whitefield SE, McCrone JT, Fitzsimmons W, Lauring AS. 2016. The mutational robustness of influenza a virus. PLoS Pathog 12:e1005856. doi: 10.1371/journal.ppat.1005856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peck KM, Chan CHS, Tanaka MM. 2015. Connecting within-host dynamics to the rate of viral molecular evolution. Virus Evol 1:vev013. doi: 10.1093/ve/vev013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Wang D, Zhang L, Sun W, Zhang Z, Chen W, Zhu A, Huang Y, Xiao F, Yao J, Gan M, Li F, Luo L, Huang X, Zhang Y, Wong S-S, Cheng X, Ji J, Ou Z, Xiao M, Li M, Li J, Ren P, Deng Z, Zhong H, Xu X, Song T, Mok CKP, Peiris M, Zhong N, Zhao J, Li Y, Li J, Zhao J. 2021. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med 13:30. doi: 10.1186/s13073-021-00847-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, Lynch J, Kidd S, Cortes N, Mori M, Williams R, Vernet G, Justice A, Green A, Nicholls SM, Ansari MA, Abeler-Dörner L, Moore CE, Peto TEA, Eyre DW, Shaw R, Simmonds P, Buck D, Todd JA, Oxford Virus Sequencing Analysis Group (OVSG), Connor TR, Ashraf S, da Silva Filipe A, Shepherd J, Thomson EC, COVID-19 Genomics UK (COG-UK) Consortium, Bonsall D, Fraser C, Golubchik T. 2021. SARS-CoV-2 within-host diversity and transmission. Science 372:eabg0821. doi: 10.1126/science.abg0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin MA, Koelle K. 2021. Comment on "genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2". Sci Transl Med 13:eabh1803. doi: 10.1126/scitranslmed.abh1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCrone JT, Lauring AS. 2018. Genetic bottlenecks in intraspecies virus transmission. Curr Opin Virol 28:20–25. doi: 10.1016/j.coviro.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang D, Wang Y, Sun W, Zhang L, Ji J, Zhang Z, Cheng X, Li Y, Xiao F, Zhu A, Zhong B, Ruan S, Li J, Ren P, Ou Z, Xiao M, Li M, Deng Z, Zhong H, Li F, Wang W-J, Zhang Y, Chen W, Zhu S, Xu X, Jin X, Zhao J, Zhong N, Zhang W, Zhao J, Li J, Xu Y. 2021. Population bottlenecks and intra-host evolution during human-to-human transmission of SARS-CoV-2. Front Med (Lausanne) 8:585358. doi: 10.3389/fmed.2021.585358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet 10:540–550. doi: 10.1038/nrg2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rockett R, Basile K, Maddocks S, Fong W, Agius JE, Johnson-Mackinnon J, Arnott A, Chandra S, Gall M, Draper J, Martinez E, Sim EM, Lee C, Ngo C, Ramsperger M, Ginn AN, Wang Q, Fennell M, Ko D, Lim HL, Gilroy N, O’Sullivan MVN, Chen S-A, Kok J, Dwyer DE, Sintchenko V. 2022. Resistance mutations in SARS-CoV-2 delta variant after sotrovimab use. N Engl J Med 386:1477–1479. doi: 10.1056/NEJMc2120219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dinis JM, Florek KR, Fatola OO, Moncla LH, Mutschler JP, Charlier OK, Meece JK, Belongia EA, Friedrich TC. 2016. Correction for Dinis et al., deep sequencing reveals potential antigenic variants at low frequencies in influenza a virus-infected humans. J Virol 90:8029. doi: 10.1128/JVI.01041-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 20:8. doi: 10.1186/s13059-018-1618-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lauring AS. 2020. Within-Host viral diversity: a window into viral evolution. Annu Rev Virol 7:63–81. doi: 10.1146/annurev-virology-010320-061642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martínez-González B, Soria ME, Vázquez-Sirvent L, Ferrer-Orta C, Lobo-Vega R, Mínguez P, de la Fuente L, Llorens C, Soriano B, Ramos-Ruíz R, Cortón M, López-Rodríguez R, García-Crespo C, Somovilla P, Durán-Pastor A, Gallego I, de Ávila AI, Delgado S, Morán F, López-Galíndez C, Gómez J, Enjuanes L, Salar-Vidal L, Esteban-Muñoz M, Esteban J, Fernández-Roblas R, Gadea I, Ayuso C, Ruíz-Hornillos J, Verdaguer N, Domingo E, Perales C. 2022. SARS-CoV-2 mutant spectra at different depth levels reveal an overwhelming abundance of low frequency mutations. Pathogens 11:662. doi: 10.3390/pathogens11060662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCrone JT, Lauring AS. 2016. Measurements of intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J Virol 90:6884–6895. doi: 10.1128/JVI.00667-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valesano AL, Rumfelt KE, Dimcheff DE, Blair CN, Fitzsimmons WJ, Petrie JG, Martin ET, Lauring AS. 2021. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog 17:e1009499. doi: 10.1371/journal.ppat.1009499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valesano AL, Taniuchi M, Fitzsimmons WJ, Islam MO, Ahmed T, Zaman K, Haque R, Wong W, Famulare M, Lauring AS. 2021. The early evolution of oral poliovirus vaccine is shaped by strong positive selection and tight transmission bottlenecks. Cell Host Microbe 29:32–43. doi: 10.1016/j.chom.2020.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koboldt DC. 2020. Best practices for variant calling in clinical sequencing. Genome Med 12:91. doi: 10.1186/s13073-020-00791-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. doi: 10.3389/fgene.2015.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stead LF, Sutton KM, Taylor GR, Quirke P, Rabbitts P. 2013. Accurately identifying low-allelic fraction variants in single samples with next-generation sequencing: applications in tumor subclone resolution. Hum Mutat 34:1432–1438. doi: 10.1002/humu.22365 [DOI] [PubMed] [Google Scholar]
- 30. Auwera G, O’Connor BD. 2020. Genomics in the cloud: Using Docker, GATK, and WDL in Terra. 1st ed. O’Reilly Media, Sebastopol, California, USA. [Google Scholar]
- 31. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrison EM. 2012. Haplotype-based variant detection from short-read sequencing. Arxiv 1207.3907V2
- 33. Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. 2019. Calling somatic SNVs and indels with mutect2. BioRxiv. doi: 10.1101/861054 [DOI]
- 34. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephens ZD, Hudson ME, Mainzer LS, Taschuk M, Weber MR, Iyer RK. 2016. Simulating next-generation sequencing datasets from empirical mutation and sequencing models. PLoS One 11:e0167047. doi: 10.1371/journal.pone.0167047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tonkin-Hill G, Martincorena I, Amato R, Lawson ARJ, Gerstung M, Johnston I, Jackson DK, Park N, Lensing SV, Quail MA, Gonçalves S, Ariani C, Spencer Chapman M, Hamilton WL, Meredith LW, Hall G, Jahun AS, Chaudhry Y, Hosmillo M, Pinckert ML, Georgana I, Yakovleva A, Caller LG, Caddy SL, Feltwell T, Khokhar FA, Houldcroft CJ, Curran MD, Parmar S, COVID-19 Genomics UK (COG-UK) Consortium, Alderton A, Nelson R, Harrison EM, Sillitoe J, Bentley SD, Barrett JC, Torok ME, Goodfellow IG, Langford C, Kwiatkowski D, Wellcome Sanger Institute COVID-19 Surveillance Team . 2021. Patterns of within-host genetic diversity in SARS-CoV-2. Elife 10:e66857. doi: 10.7554/eLife.66857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, Birol I. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kellman BP, Baghdassarian HM, Pramparo T, Shamie I, Gazestani V, Begzati A, Li S, Nalabolu S, Murray S, Lopez L, Pierce K, Courchesne E, Lewis NE. 2021. Multiple freeze-thaw cycles lead to a loss of consistency in poly(A)-enriched RNA sequencing. BMC Genomics 22:69. doi: 10.1186/s12864-021-07381-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li L, Li X, Guo Z, Wang Z, Zhang K, Li C, Wang C, Zhang S. 2020. Influence of storage conditions on SARS-CoV-2 nucleic acid detection in throat swabs. J Infect Dis 222:203–205. doi: 10.1093/infdis/jiaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao L, Illingworth CJR. 2019. Measurements of Intrahost viral diversity require an unbiased diversity metric. Virus Evol 5:vey041. doi: 10.1093/ve/vey041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental setup of simulated and synthetic data generation.
Variant caller performance using default and custom parameters on simulated data.
Effect of coverage cutoffs and sequencing replicates on false discovery rate and false negative rate in synthetic and simulated data.
Effect of frequency cutoffs and sequencing replicates on false discovery rate and false negative rate in simulated data.
Effect of sequencing replicates and genome location on the accuracy of allele frequency estimation.
Quantification of majority and minority variants identified in data from SARS-CoV-2 clinical specimens.
Reproducibility of minority variants across sequencing replicates from the Tonkin-Hill et al. 2021 sequencing data set.
List of variant callers and parameters; list of diagnostic samples.
Details on some of methods.
Data Availability Statement
Synthetic influenza data (Bioproject PRJNA865369) and SARS-CoV-2 data from clinical samples are available in NCBI GenBank and SRA (Bioproject PRJNA857712). Accession IDs can be found in Table S2. All downstream analysis files are available at https://github.com/GhedinSGS/Optimized-Quantification-of-Intrahost-Viral-Diversity. R functions for performing SNV analysis and generating plots were compiled into an R package, vivaldi (Viral Variant Location and Diversity), available at https://cran.r-project.org/web/packages/vivaldi/index.html.