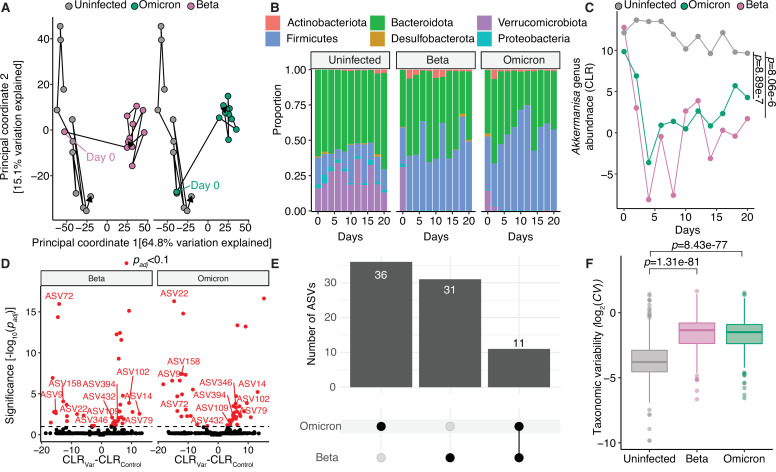
Fig 5.
Mild severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in wild-type mice result in destabilization to the gut microbiota and loss of Akkermansia. (A) Principal coordinate analysis of C57BL/6J mice comparing communities after Beta and Omicron variant infection are shown relative to uninfected mice. (B) Phylum-level taxonomic summaries for uninfected or Beta- or Omicron-infected mice sampled from days 0 to 20 after inoculation. (C) Akkermansia genus, Centered Log Ratio (CLR) for each variant plotted against day of sampling post-inoculation. P values, two-way analysis of variance comparing each variant to the uninfected group. (D–E) A linear mixed effects model was created for each variant. (D) Volcano plot of padj versus difference in CLR abundance. (E) Differentially abundant amplicon sequence variants (ASVs) shared/distinct between variants. Overlapping ASVs in (E) are annotated in (D). (F) Taxonomic variability is plotted comparing uninfected and Beta- and Omicron-infected mice. n = 15 mice with five mice per group in three cages, and 33 total longitudinally collected samples.