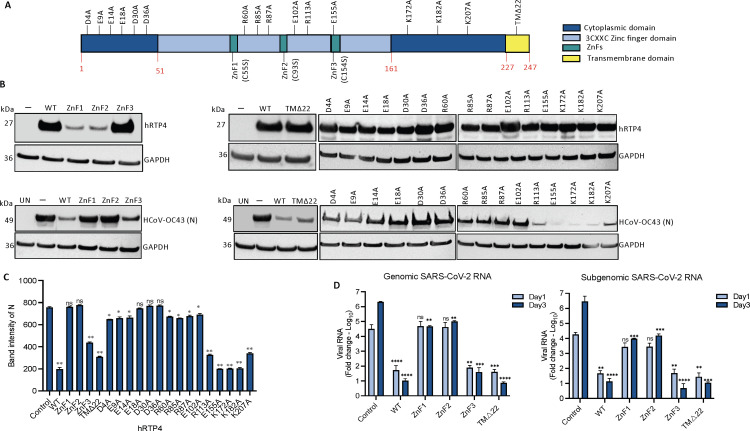
FIG 4 .
The N-terminal domain of hRTP4 is required for its antiviral activity. (A) The domain structure of RTP4 is diagrammed. (B) CHME3 cells were transfected with expression vectors for hRTP4 mutants ZnF1, ZnF2, and ZnF3, a 22 amino acid C-terminal deletion TMΔ22 and the indicated charged residue point mutants. Wild-type (WT) hRTP4 and empty vector served as controls. Expression levels were quantified on an immunoblot probed with anti-FLAG and GAPDH antibodies (top). The transfected cells were infected with HCoV-OC43 at an MOI of 0.5. After 48 h, the HCoV-OC43 N protein was quantified by immunoblot analysis with GAPDH as a loading control (bottom). (C) Band intensities from the immunoblot analysis for the HCoV-OC43 N protein expression levels are plotted. The experiment was repeated with similar results. All the blots from independent experiments were analyzed at the same exposure time. (D) ACE2.CHME3 cells were infected with SARS-CoV-2 at an MOI of 0.1. Viral genomic and subgenomic RNAs were quantified by qRT-PCR at 1 and 3 dpi. The data are normalized to GAPDH and expressed as fold-change relative to the uninfected cells. All data are the averages of three biological replicates (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).