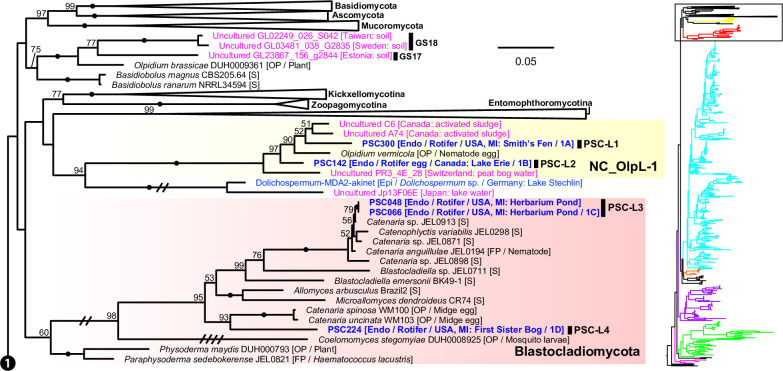
Fig 6.
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including Ascomycota, Basidiomycota, Mucoromycota, Entomopthoromycotina, Kickxellomycotina, Zoopagomycotina, Blastocladiomycota, and the NC_OlpL-1 clade. ML bootstrap values higher than 50% were shown on each branch. Black dots on branches indicate 100% bootstrap value. Double and quadruple slashes on branches indicate that length is reduced by half and quarter, respectively. Cultured fungi are labeled in black; saprotrophs are indicated as [S], and obligate [OP] and facultative [FP] parasites are indicated as [O(F)P / its host]. Single cells isolated in this study are labeled in bold blue and previously published sequences of single cells are labeled in blue; annotations are indicated as [Endo (endobiotic) or Epi (epibiotic) / host / isolation source / figure number if available]. Published environmental DNA sequences are labeled in pink and PacBio OTU sequences in this study are labeled in bold red; source of each sequence is described in parentheses.