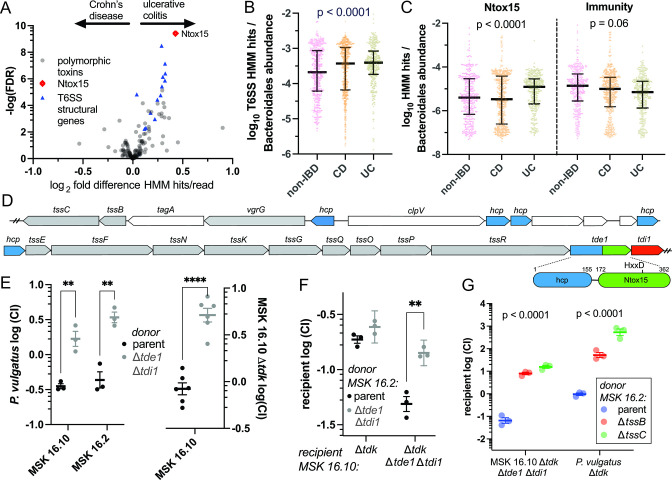
FIG 1.
Ntox15 domains enriched in IBD metagenomes mediate T6SS-dependent interbacterial antagonism among Bacteroidales. Metagenomic sequencing reads with similarity to Bacteroidales T6SS, Ntox15 domains, and immunity proteins were detected with hidden Markov models [HMMer (33)]. (A) T6SSiii structural genes and Ntox15 domain homologs are enriched in fecal metagenomes from patients with UC compared to CD (32). False discovery rate adjustment for multiple comparisons was with the Benjamini–Hochberg method. (B, C) Aggregated T6SS structural genes and Ntox15 homologs, but not the associated immunity are enriched in UC over CD and non-IBD controls after correction for relative Bacteroidales abundance. P-value reflects Kruskal–Wallis test. (D) A gene structure diagram of a T6SS-encoding locus that is identical in several genetically diverse Bacteroidales isolates from a single human donor. In addition to other T6SS structural genes (gray), there are five hcp genes (blue), including one fused with a C-terminal Ntox15 domain (tde1, green) and an immediately adjacent immunity gene (tdi1, red). An HxxD motif is conserved at the putative active site, predicted to confer nuclease activity. (E) In competitive growth experiments with P. vulgatus ATCC 8482, deletion of tde1 and tdi1 from MSK 16.10 or MSK 16.2 confers reduced relative fitness. Effector/immunity deletion is also a competitive disadvantage relative to the isogenic parental strain. Thymidine kinase (tdk) is deleted to confer resistance to the selection agent floxuridine (FUdR). (F) tde1/tdi1 mediate competition between MSK 16.10 and MSK 16.2, isolates from a single human host. Statistical indicators reflect Student’s t-test: ** P < 0.01, *** P < 0.001. (G) Tde1-dependent antagonism requires structural sheath proteins TssB and TssC. P-values reflect analysis of variance (ANOVA) tests for each recipient.