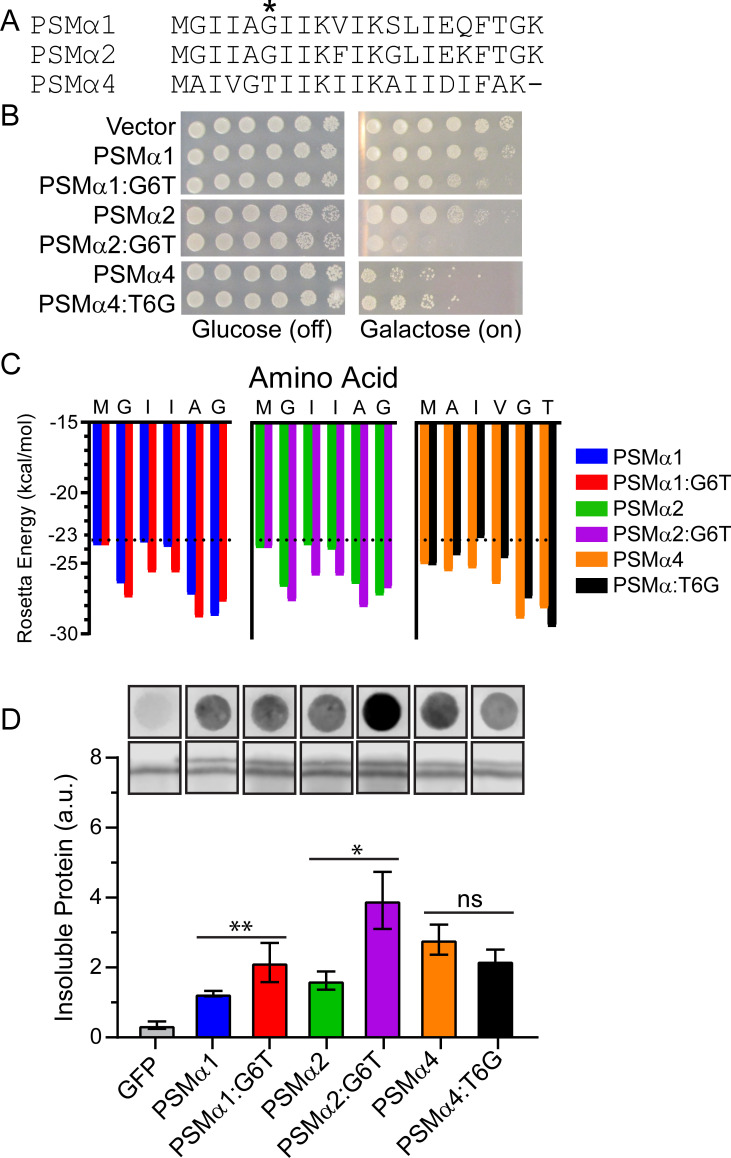
Fig 3.
Missense mutations of PSMα residue 6 drive propensity for aggregation, toxicity, and solubility. (A) Sequence alignment PSMα1, PSMα2, and PSMα4 indicates lack of conservation at residue 6. (B) BYΔhsp104 yeast were transformed with the indicated 423GAL-PSMα-GFP plasmid or 423GAL-GFP control. Strains were serially diluted 5-fold and spotted on glucose (off) or galactose (on) media. (C) Analysis of the sequences by ZipperDB (50). Steric zippers are predicted to form when the Rosetta energy of a hexapeptide is below the threshold of −23 kcal/mol, which is denoted by a hashed line. (D) Strains from B were induced for 5 h, spheroplasted, and lysed. Extracts were then passed over a nonbinding cellulose acetate (CA) membrane. Membranes were then probed using an anti-GFP antibody. Representative results from three replicates are shown (top row). Extracts were also probed via immunoblotting to assess total GFP (second row). The ratio of aggregated protein (top row, bound to CA) to total PSMα-GFP protein (second row, from immunoblotting) was calculated (bottom). Aggregation of each WT PSMα protein was compared to its respective mutant using two-tailed t-tests. (N ≥ 6, bars show mean ± SEM, *P < 0.05, and **P < 0.01). Note that the shown filter retention assay and immunoblotting samples were all run on a single membrane in a randomized order, with a single representative trial shown. Lanes are cropped and re-ordered for presentation purposes.