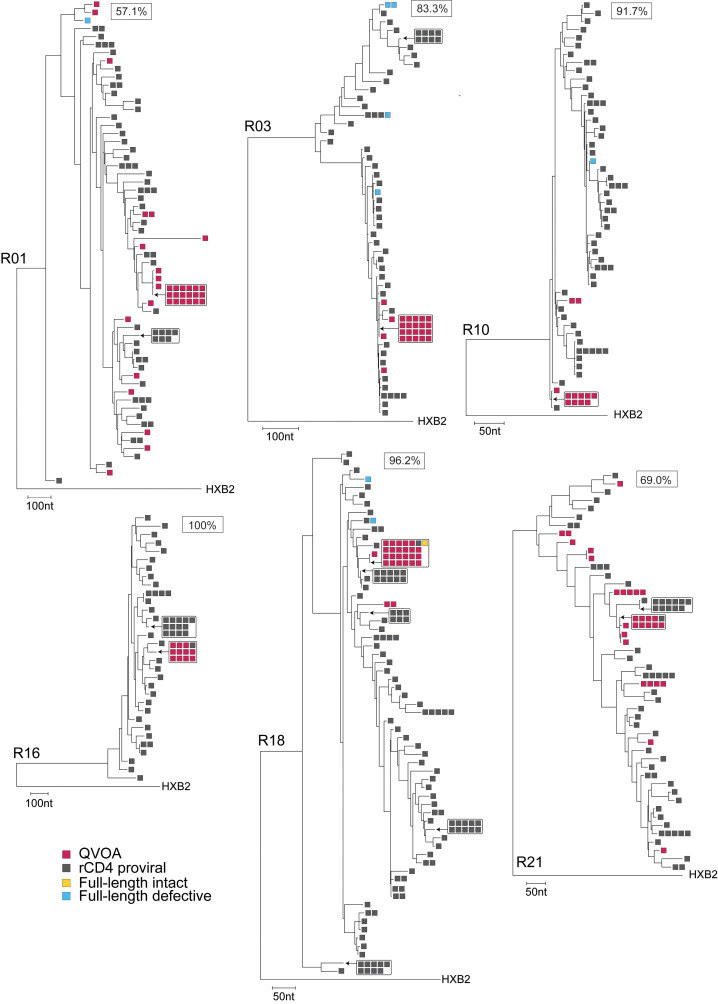
Figure 5. Large expanded CD4+ T cell clones dominate the latent reservoir of PWH on very long-term ART.
Maximum-likelihood phylogenetic trees of env sequences for 6 representative participants (R01, R03, R10, R16, R18, and R21), each rooted using HXB2, are shown. Single-genome sequencing of env was performed on cDNA reverse-transcribed from viral RNA extracted from supernatants of QVOA wells scored positive for viral outgrowth (red) or on proviral DNA from resting CD4+ T cells (rCD4, gray). Near-full-length genome sequences were obtained using the same proviral DNA and are annotated as intact (yellow) or defective (blue). Genetic distance is represented by the scale in nucleotides. A measure of the clonality of QVOA isolates (see article text) is indicated as a boxed value for each participant.