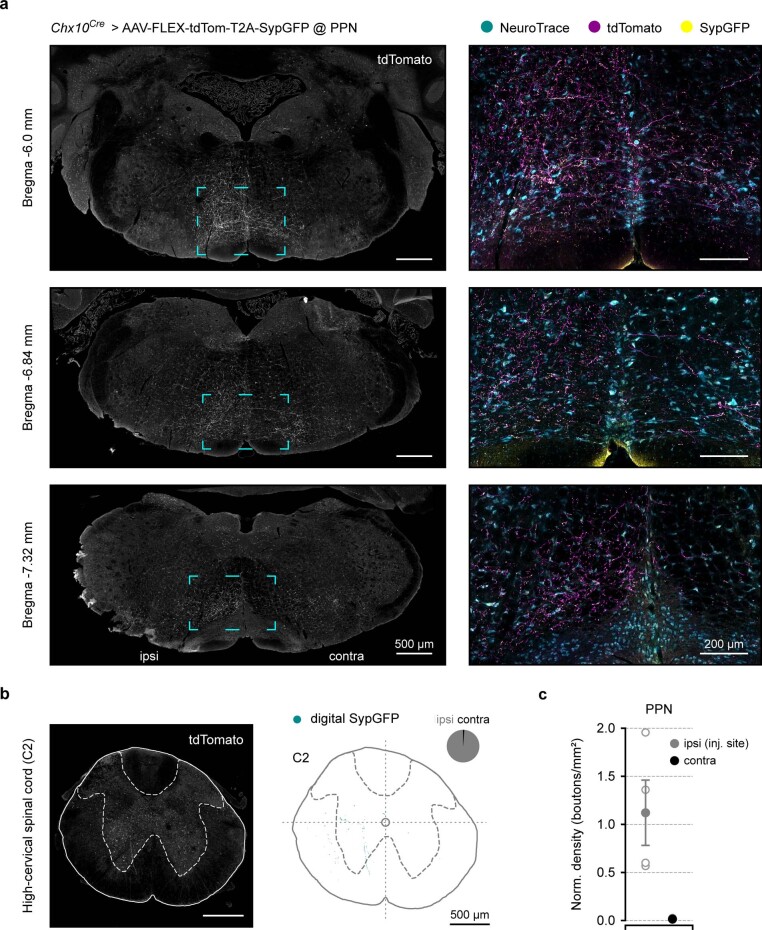
Extended Data Fig. 9. Representative images of the anterograde tracing and lack of projections to the contralateral PPN.
a, Example confocal photomicrographs at three different brainstem levels (coronal plane) from a Chx10Cre mouse unilaterally injected in the PPN with the anterograde tracer AAV-FLEX-tdTom-T2A-SypGFP to reveal the projection pattern of Chx10-PPN neurons. Left, single-channel images (grayscale) of brainstem coronal sections (excluding cerebellum) at −6.0, −6.84, and −7.32 mm from bregma. White fluorescence signal corresponds to tdTomato, which fills the cell-bodies and fibers of the transfected Chx10-PPN neurons. The whole tissue and associated anatomical landmarks are visible through background auto-fluorescence. Dashed turquoise squares delineate the magnified areas to the right. Scale bars 500 µm. Right, multi-channel composite images of the magnified areas on the left containing a fluorescent Nissl stain (NeuroTrace, cyan), fibers from the transfected Chx10-PPN neurons (tdTomato, magenta), and their synaptic boutons (SypGFP, yellow). Scale bars 200 µm. b, Left, example single-channel (grayscale) confocal photomicrograph of a coronal spinal cord section at C2 level (high cervical spinal cord) where white fluorescence signal corresponds to tdTomato as in a, showing very sparse labelling. Right, digital reconstruction of putative synaptic bouton positions (digital SypGFP, cyan) from the example cervical spinal cord image on the left. The pie chart depicts the percentage of boutons ipsilateral (ipsi, gray) or contralateral (contra, black) to the injection site. Scale bars 500 µm. c, Normalized bouton density (boutons/mm2) at the injection site (PPN, ipsilateral, gray), compared to the contralateral PPN (black). Chx10-PPN neurons do not project to the contralateral PPN. Solid circles (ipsi, gray; contra, black) are the group average (all mice), error bars represent s.e.m., and hollow circles individual mice (N = 4 mice).