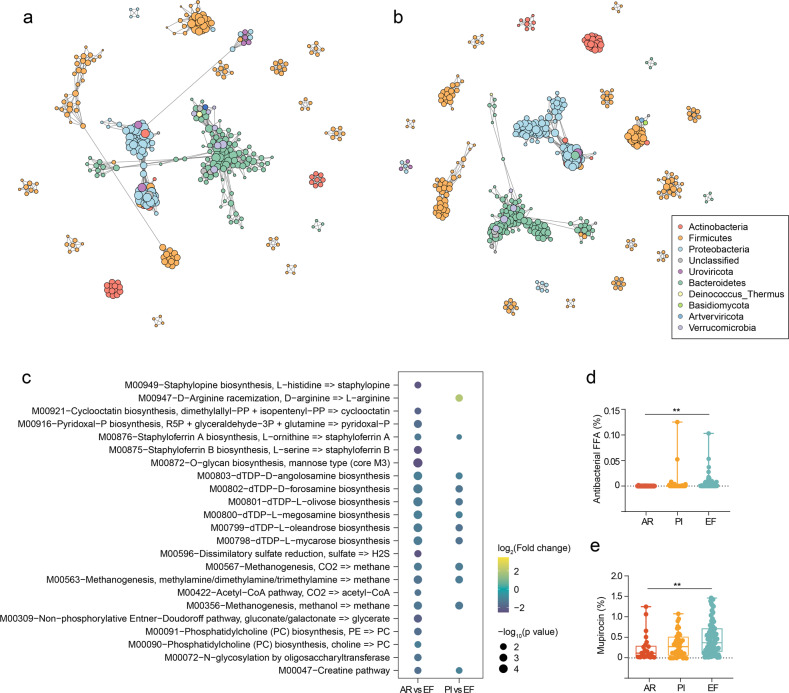
Fig. 3.
The microbial association network for a allograft disorder and b EF specimens. The co-abundance correlation between the species was calculated using SparCC correlation. All significant correlations with BH-adjusted p < 0.05 were included. Each node represents a microbial species. Edges between nodes represent correlations. c The significant changed Kyoto Encyclopedia of Genes and Genomes (KEGG) modules in AR vs EF and PI vs EF. The significantly differential modules were defined by false discovery rate less than 0.1 in Wilcox Rank-Sum Test and fold change between two groups more than 2. d Antibacterial FFA and e mupirocin-related genes in gut microbiota of three groups. The center line of the box represents the median, and the box bounds represents the inter-quartile range. The whiskers span 1.5-fold the inter-quartile range. The comparison among the groups was tested by Kruskal–Wallis test. **p < 0.01