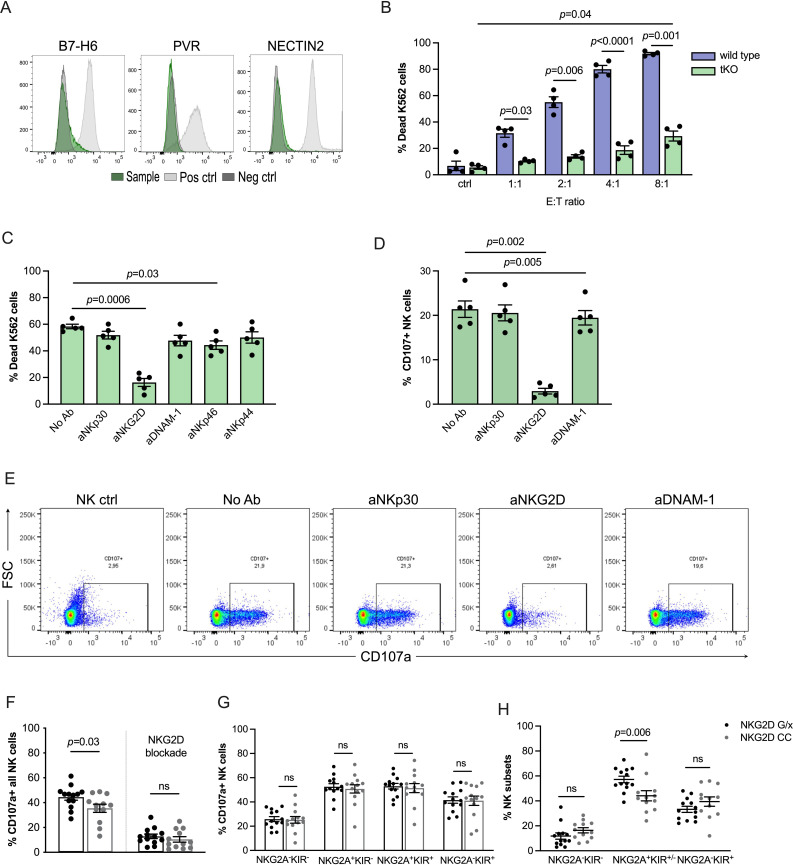
Figure 2.
NK cell function based on NKG2D genotypes using an NKG2D-dependent model. (A) Flow cytometry staining of triple KO (tKO) K562 cells showing successful knockout of ligand genes. (B) Cytotoxicity assay of healthy donor resting NK cells against wt K562 and tKO K562 cells at various effector:target (E/T) ratios (n=4). (C) Cytotoxicity assay of polyclonally activated NK cells from healthy donors against triple knockout K562 at 8:1 E/T ratio, in the presence or absence of indicated NK cell receptor blockade (n=5). (D, E) Degranulation assay of healthy donor resting NK cells against tKO K562 combined with NK cell receptor blockade (n=5) with representative dot plots. (F) Degranulation assay of CD56+NK cells from healthy donors with NKG2D G/x or CC, preactivated with IL-15 overnight and cocultured on next day with tKO K562 for 4 hours in the presence or absence of NKG2D blocking antibody (G/x n=13 and CC n=12). (G) Degranulation response in different NKG2A+/− and KIR+/− NK cell subsets in healthy donors with G/x (n=13) or CC (n=12) genotypes. (H) Frequency of different NK cell subsets in healthy donors according to NKG2D SNP G/x (n=13) and CC (n=12). Error bars represent SEM. NK, natural killer; SNP, single nucleotide polymorphism.