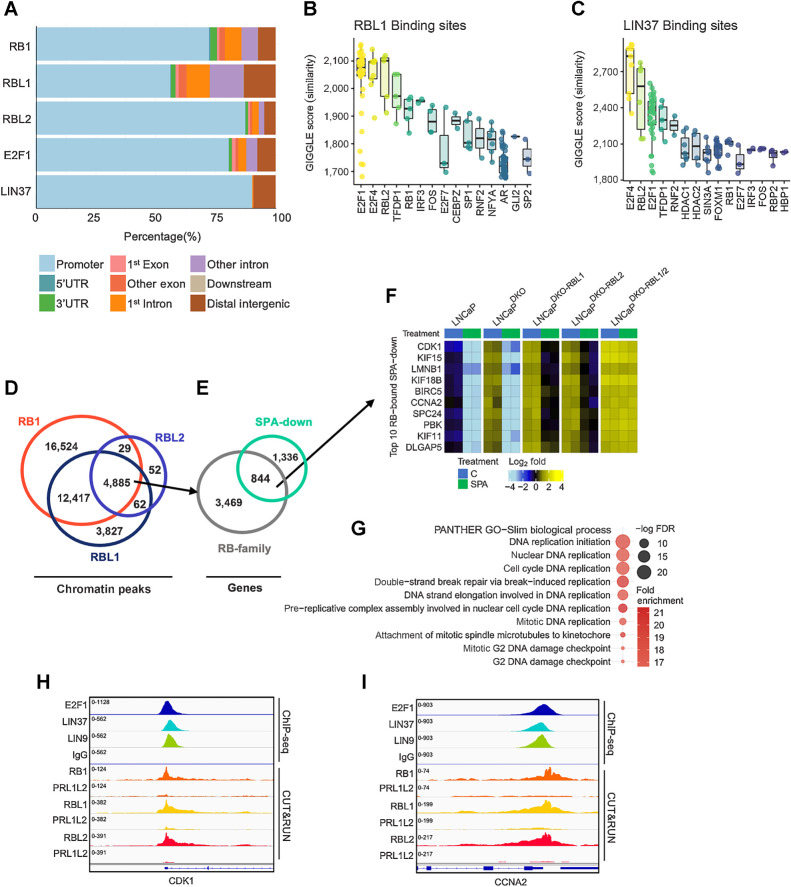
Figure 3.
Androgen-repressed cell-cycle genes bound by the DREAM complex. A, RB-family, E2F1, and LIN37 genomic binding sites determined by ChIP-seq from LNCaP cells and mapped to annotated regions. B, Boxplots of GIGGLE similarity scores for RBL1 ChIP-seq from LNCaP cells. C, Boxplots of GIGGLE similarity scores for LIN37 ChIP-seq from LNCaP cells. D, Overlapping CUT&RUN peaks bound by RB1 LNCaP cells, RBL1 in LNCaPDKO-RBL2 cells, and RBL2 LNCaPDKO-RBL1 cells. E, RB-family co-bound sites mapped to gene promoters and overlapped with SPA-repressed genes in LNCaPDKO cells (FDR < 0.05). F, Heatmap of mean-centered log2(FPKM) RNA-seq values for the top 10 androgen-repressed genes bound by all three RB-family members in LNCaP and isogenic gene knockout sublines. G, GO enrichment analysis on androgen repressed genes co-bound by RB-family members. H and I, Chromatin peaks for ChIP-seq datasets for E2F1 (LNCaPDKO), LIN37 (LNCaP), LIN9 (LNCaP), and IgG (LNCaPDKO); and for CUT&RUN peaks for RB1 (LNCaP), RBL1 (LNCaPDKO-RBL2), RBL2 (LNCaPDKO-RBL1), and LNCaP DKO-RBL1/2 (PRL1L2) as a negative control for each antibody on the promoters for SPA-repressed genes CDK1 (H) and CCNA2 (I).