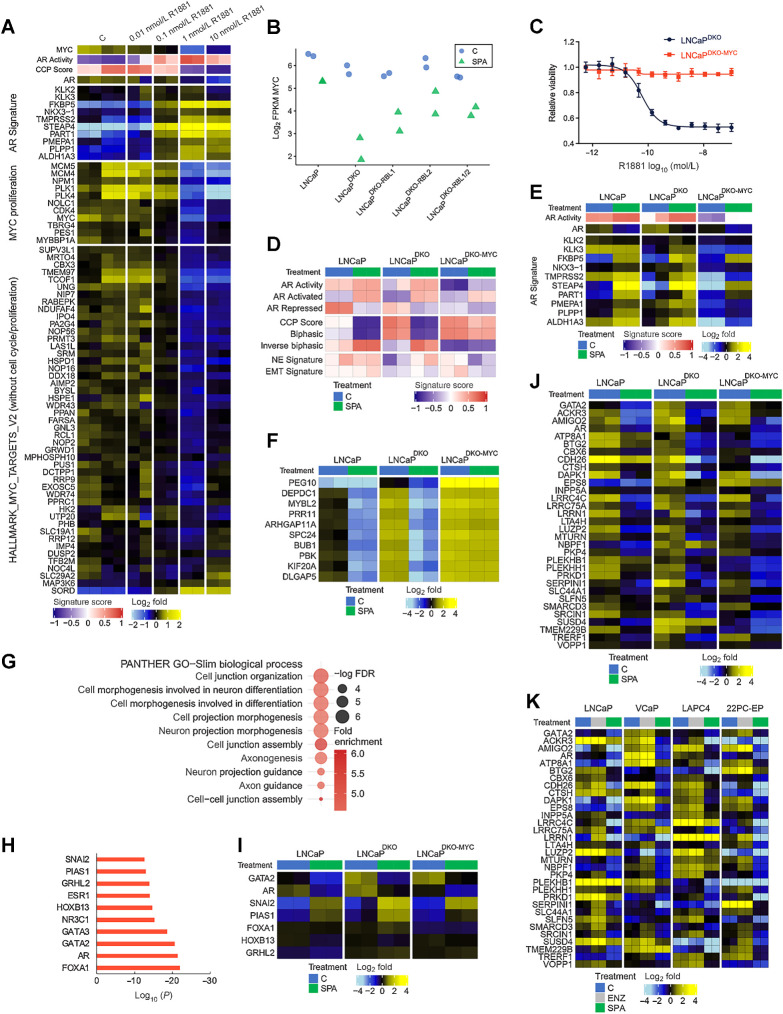
Figure 6.
MYC expression is a key determinant of response to high-dose androgen. A, Heatmaps of RNA-seq mean-centered log2(FPKM) gene expression values and AR and MYC-regulated genes in LNCaP cells treated with a dose-range of R1881. C, vehicle control. B, RNA-seq log2(FPKM) expression of MYC is plotted for LNCaP and isogenic knockout lines treated with SPA for 48 hours or vehicle control. C, Dose–response curve to R1881 for LNCaPDKO cells with or without ectopic MYC overexpression. D, Heatmap of molecular signature (GSVA) scores derived from RNA-seq data from LNCaPDKO cells with or without ectopic expression of MYC treated with SPA for 48 hours. CCP, cell-cycle proliferation; EMT, epithelial-to-mesenchymal transition; NE, neuroendocrine. E, Heatmap of mean-centered log2(FPKM) RNA-seq values for the AR activity signature genes determined from RNA-seq data from SPA or vehicle control in LNCaP, LNCaPDKO, and LNCaPDKO-MYC cells. GSVA scores and treatment groups are shown at the top of plot and colored according to legends at the bottom. LNCaP and LNCaP-DKO data are replotted from Fig. 1. F, Heatmap of mean-centered log2(FPKM) RNA-seq values for the top 10 genes differentially regulated by MYC overexpression in LNCaPDKO cells treated with SPA. G, GO enrichment analysis of MYC/E2F-independent AR-repressed genes in LNCaPDKO cells. H, Top motifs predicted to regulate MYC/E2F-independent AR-repressed genes by LISA. I, Heatmap of mean-centered log2(FPKM) RNA-seq values for predicted regulators of MYC/E2F-independent AR-repressed genes. J, Heatmap of mean-centered log2(FPKM) RNA-seq values for consensus MYC/E2F-independent AR-repressed genes (AR-repressed signature genes repressed by SPA in LNCaPDKO-MYC cells). LNCaP and LNCaP-DKO data are replotted from Fig. 1. K, Heatmap of mean-centered log2(FPKM) RNA-seq values for consensus MYC/E2F-independent AR-repressed genes comparing SPA versus vehicle control in LNCaP, VCaP, LAPC4, and 22PC-EP cell lines.