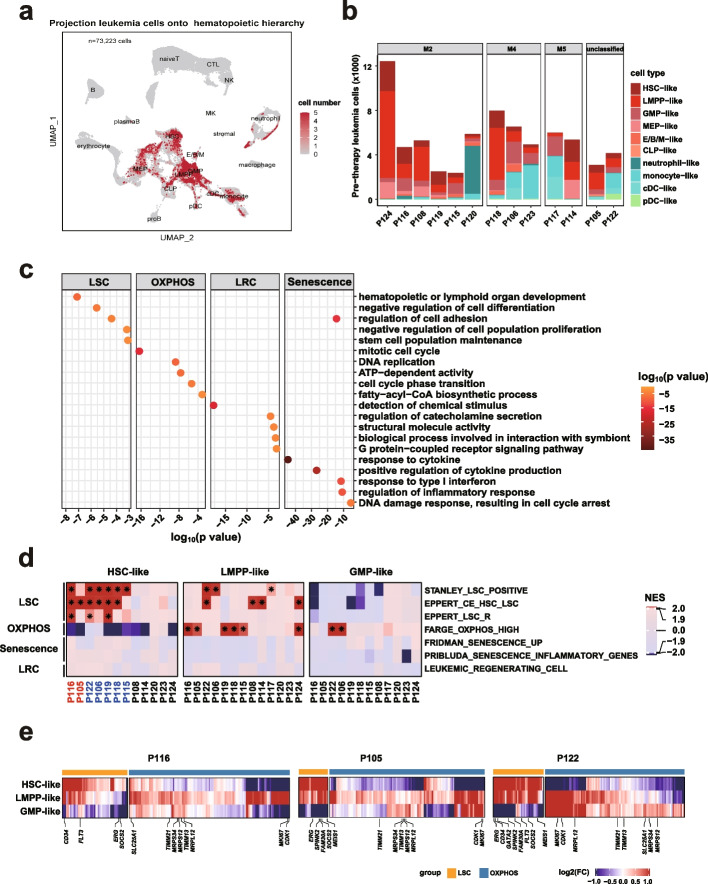
Fig. 3.
LSC and OXPHOS signatures were prevalent in leukemic stem and progenitor populations. a UMAP plot illustrating the projection of transcriptionally predicted leukemic cells from thirteen AML patients onto the normal hematopoietic hierarchy, based on transcriptomic similarity to normal cells. Projected cells are highlighted, with shading indicating the frequency of being projected. b Bar plot showing the cell counts of pre-therapy leukemia cell populations in each AML patient. c Dot plot displaying pathways enriched by four known chemoresistance-related expression signatures derived from mouse model studies, with colors representing enrichment p values. d Heatmap depicting the GSEA results of the four expression signatures in panel c for each HSPC-like population compared to all other leukemic populations within each patient before therapy. Colors represent NES values obtained from GSEA analysis, and an asterisk denotes both NES > 1.9 and FDR < 0.001. Patient code colors indicate resistant (red) and sensitive (blue) cases. e Heatmaps showing expression fold changes (FC) of core enriched genes (columns) contributing to LSC and OXPHOS signatures in each HSPC-like population (rows) compared to all other leukemic populations in three representative patients (P116, P105, and P122). Core enriched genes are identified from GSEA results and those related to cell stemness and metabolism are indicated