Figure 4. ME2 responds to fumarate by modulating mitoribosome assembly.
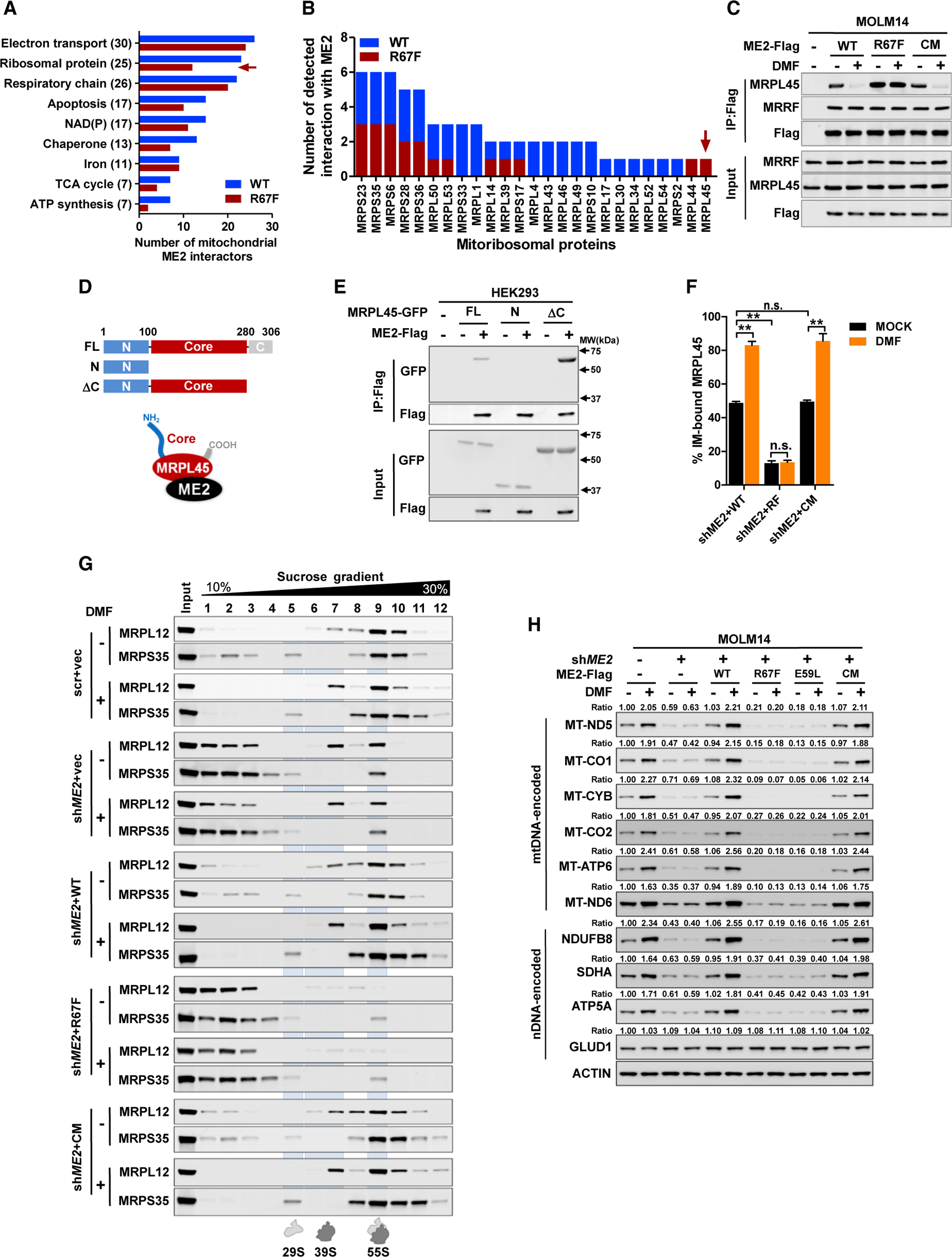
(A) ME2 interactors were functionally grouped; the number on the y axis indicates total number of wild-type or mutant ME2-binding proteins.
(B) ME2-interacting proteins were identified by pull-down mass spectrometry in three independent experiments. The number of detected interactions of ME2 (wild-type and R67F mutant) with mitoribosomal proteins was determined.
(C) MOLM14 cells expressing ME2-Flag and its mutants were treated with DMF. The interaction of ME2 with MRPL45 and MRRF was determined.
(D and E) GFP-tagged full-length MRPL45 (FL) and its mutants (N and ΔC) (D) were co-expressed with ME2-Flag to determine their association (E).
(F) ME2-knockdown and re-expression MOLM14 cells were treated with DMF. Isolated mitochondria were fractionated to determine MRPL45 localization. (G and H) ME2-knockdown and re-expression MOLM14 cells were treated with DMF for 24 h. Isolated mitochondria were loaded on a sucrose gradient to fractionate mitoribosome (G). mtDNA and nDNA-encoded proteins were determined (H).
All data are presented as mean ± SEM from three independent experiments. **p < 0.01; n.s. indicates not significant. See also Figure S4.
