The HBZ viral oncogene expressed in HTLV-1–infected T cells upregulates TAp73 by both RNA and protein forms, modulating lactate metabolism and epigenetic status to accelerate oncogenesis of adult T-cell leukemia/lymphoma.
Abstract
Acceleration of glycolysis is a common trait of cancer. A key metabolite, lactate, is typically secreted from cancer cells because its accumulation is toxic. Here, we report that a viral oncogene, HTLV-1 bZIP factor (HBZ), bimodally upregulates TAp73 to promote lactate excretion from adult T-cell leukemia-lymphoma (ATL) cells. HBZ protein binds to EZH2 and reduces its occupancy of the TAp73 promoter. Meanwhile, HBZ RNA activates TAp73 transcription via the BATF3-IRF4 machinery. TAp73 upregulates the lactate transporters MCT1 and MCT4. Inactivation of TAp73 leads to intracellular accumulation of lactate, inducing cell death in ATL cells. Furthermore, TAp73 knockout diminishes the development of inflammation in HBZ-transgenic mice. An MCT1/4 inhibitor, syrosingopine, decreases the growth of ATL cells in vitro and in vivo. MCT1/4 expression is positively correlated with TAp73 in many cancers, and MCT1/4 upregulation is associated with dismal prognosis. Activation of the TAp73–MCT1/4 pathway could be a common mechanism contributing to cancer metabolism.
Significance:
An antisense gene encoded in HTLV-1, HBZ, reprograms lactate metabolism and epigenetic modification by inducing TAp73 in virus-positive leukemic cells. A positive correlation between TAp73 and its target genes is also observed in many other cancer cells, suggesting that this is a common mechanism for cellular oncogenesis.
This article is featured in Selected Articles from This Issue, p. 337
INTRODUCTION
The hallmarks of cancer, a framework for understanding the common features of diverse human malignant tumors, include “deregulating cellular metabolism” and “epigenetic reprogramming” (1, 2). The Warburg effect is a well-known energy-generation phenomenon associated with cancer metabolism, by which the rate of glucose uptake, glycolysis, and consequent production of lactate are drastically increased in cancer cells even under aerobic conditions (3, 4). Epigenetic alterations, such as CpG DNA hyper- or hypomethylation and aberrant histone modifications, dysregulate chromatin structure and gene expression to promote oncogenesis. Although many types of malignancy share these hallmarks and enabling characteristics, the pathogenesis of each tumor is complex. To elucidate the molecular mechanisms for oncogenesis of each malignant disease, one must consider the characters of each cell type and any associated exogenous factors, such as pathogens.
Human T-cell leukemia virus type 1 (HTLV-1) is an oncogenic retrovirus that mainly infects CD4+ T cells and causes a fatal malignancy called adult T-cell leukemia-lymphoma (ATL). As viral replication of HTLV-1 is generally suppressed in vivo, persistent infection is established mainly by clonal proliferation of infected cells. Among the viral genes, only HTLV-1 bZIP factor (HBZ), which is encoded in the minus strand of the provirus, is conserved and expressed in all ATL cases. Knockdown of HBZ reduces the proliferation of ATL cell lines (5), and transgenic mice that express HBZ in CD4+ T cells (HBZ-Tg mice) develop systemic inflammation and T-cell lymphoma (6), suggesting that HBZ contributes to the pathogenesis of HTLV-1. Additionally, HBZ protein modulates the immunophenotypes of expressing cells toward a regulatory T cell (Treg)–like phenotype by inducing Treg-associated genes such as Foxp3 (7–9). A fascinating feature of the HBZ gene is that its transcript not only encodes the HBZ protein but also acts like long noncoding RNA (lncRNA; refs. 5, 10). Previous studies using microarray analysis suggested that HBZ RNA promotes the expansion of CD4+ T cells by inducing cell cycle– and apoptosis-related genes (5, 10). Thus, HBZ plays critical roles in the clonal expansion of infected cells and the development of ATL by using both its coding and noncoding functions.
In this study, we show that HBZ induces reprogramming of lactate metabolism by upregulating the cellular transcription factor TAp73, and that HBZ also modulates the function of the enhancer of zeste homolog 2 protein (EZH2), leading to widespread epigenetic alterations. HBZ RNA and HBZ protein each activate the promoter of TAp73 by different mechanisms, and TAp73 consequently induces expression of MCT1 and MCT4, which act as lactate exporters to optimally excrete lactate created in ATL cells. EZH2 is one of the subunits of polycomb repressive complex 2 (PRC2), which functions as a methyltransferase to repress transcription by trimethylating histone H3 at lysine 27 (H3K27me3; ref. 11). Gain-of-function mutations of EZH2 are found in several cancer types, whereas ATL is shown to have high expression levels of unmutated EZH2 (12–14). Here we show that HBZ protein binds to EZH2 and changes the genome-wide distribution of H3K27me3, resulting in high expression of TAp73 as well as other widespread changes to the transcriptome. In addition, TAp73 further activates the transcription of EZH2 in ATL cells.
RESULTS
Both HBZ Protein and RNA Upregulate TP73
Both HBZ protein and HBZ RNA modulate the cellular transcriptome through dysregulation of the promoter activity of target genes (10, 15). To comprehensively analyze the target genes of HBZ protein and RNA, we began by conducting RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin sequencing (ATAC-seq) of primary murine CD4+ T cells expressing wild-type (WT) HBZ, mutant HBZ that can act only in protein form, or mutant HBZ that can act only in RNA form (Fig. 1A). Although the modes of action of HBZ protein and RNA were expected to be different, we found that all molecular forms of HBZ (WT, protein mutant, and RNA mutant) had similar transcriptional profiles in some gene clusters (Fig. 1B). Comparative analysis showed that 467 upregulated genes (Fig. 1C, left) and 371 altered opened-chromatin regions (Fig. 1C, right) were affected by all forms of HBZ. Indeed, Ccr4 and Tigit are already known to be common targets, and the present results confirmed this to be the case (Fig. 1D; refs. 7, 8, 16). Gene set enrichment analysis (GSEA) also supported the observation that both HBZ protein and HBZ RNA target similar gene sets, such as TP53- or KRAS-related gene sets (Supplementary Fig. S1A–S1C).
Figure 1.
HBZ protein and RNA both upregulate TP73. A, Schematic diagram for retroviral transfer of HBZ into primary mouse CD4+ T cells. Each construct encodes WT HBZ, HBZ RNA (ATG is converted to TTG) or HBZ protein (SM, silent mutations; ref. 5). Created with BioRender.com. B, A heat map of the top 500 differentially expressed genes in HBZ WT and its mutants compared with the vector, calculated from RNA-seq data. C, Shared transcriptomes (left; the number of genes) and open chromatin regions (right) associated with HBZ WT and its mutants are shown in Venn diagrams. D, Volcano plots of differentially expressed genes. Fold change and adjusted P value (Padj) are plotted for genes that are upreulgated (yellow) or downregulated (blue) compared with the vector. E, TP73 gene maps (upper, mouse; lower, human) depicting the major two isoforms, TAp73 and DNp73, and their promoters. F, Transcripts per million (TPM) of TAp73 in the transduced cells (n = 3). G, A volcano plot resulting from RNA-seq that compares the CD4+ T cells of HBZ-Tg mice with those of WT mice (n = 3). H, TPM of TAp73 in HBZ-Tg and WT mouse CD4+ T cells (n = 3). I–K, TP73 expression in ATL. A volcano plot resulting from RNA-seq compares CD4+ T cells of ATL patients (n = 7) with those of healthy donors (n = 10; I). mRNA expression of TAp73 (J) and DNp73 (K) by RT-qPCR in CD4+ T cells of ATL patients (acute type, n = 28; lymphoma type, n = 5; chronic type, n = 7) and healthy donors (hCD4; n = 8). Results are plotted as mean ± SD, using one-way ANOVA followed by the post hoc Steel test (J and K). ***, P < 0.001; ns, not significant.
Among the common target genes, we focused on Trp73 because it was highly expressed in cells containing any of the three HBZ constructs (Fig. 1D). Because it has alternative promoters, Trp73 encodes two major isoforms: TAp73, a full-length protein with a transactivation (TA) domain, and DNp73, an N-terminal truncated protein that lacks the TA domain (Fig. 1E; ref. 17). Among the two isoforms, TAp73 was upregulated by HBZ (Fig. 1F), whereas DNp73 was not (at least in mouse cells, see below; Supplementary Fig. S1D). Similarly, CD4+ T cells of HBZ-Tg mice, in which HBZ expression is driven by the CD4 promoter (6, 18), showed upregulation of TAp73 but not of DNp73 (Fig. 1G and H; Supplementary Fig. S1E). We next evaluated clinical samples from patients with ATL, and we found that both TAp73 and DNp73 were upregulated in human ATL cells (Fig. 1I–K). Moreover, the upregulation of both transcripts was associated with acute-type ATL, which is the most aggressive clinical subtype (Fig. 1J and K). Altogether, these observations suggest that the TP73 gene, particularly TAp73, is a target of both HBZ protein and HBZ RNA.
TAp73 has been thought to exert cancer-suppressive functions due to its proapoptotic effect and genomic integrity maintenance (19, 20). It, thus, seemed counterintuitive that TAp73 would be upregulated in ATL cells. We, therefore, analyzed The Cancer Genome Atlas (TCGA) RNA-seq data sets to assess transcriptional profiles of the TP73 gene in a variety of cancer cells. Surprisingly, the vast majority of registered cancer cells have enhanced expression of TP73, and in particular they have a greater enhancement of expression of TAp73 than of DNp73 (Supplementary Fig. S1F–S1H). Further exploration of any potential oncogenic effects of TAp73 in ATL cells could, therefore, elucidate possible oncogenic mechanisms in a broader class of cancer types.
HBZ Protein Complexes with EZH2 and Reduces Its Recruitment to the TAp73 Promoter without Inhibiting Its Methyltransferase Activity
To investigate the mechanisms for TAp73 induction by HBZ protein and RNA, we next analyzed the genetic elements in the promoter of TAp73 using data sets of chromatin immunoprecipitation sequencing (ChIP-seq) provided by the Encyclopedia of DNA Elements (ENCODE; refs. 21, 22). Around the TAp73 promoter (P1 in Fig. 1E), the target regions of two components of PRC2 (EZH2 and SUZ12) were significantly enriched (Supplementary Fig. S2A).
Our RNA-seq results also showed that HBZ WT and protein, but not RNA, enhanced the same gene sets (PRC2_EZH2_UP.V1_UP) that are upregulated when EZH2 is knocked down (Fig. 2A; Supplementary Fig. S1A), suggesting that HBZ protein might be involved in epigenetic regulation through EZH2. Indeed, immunoprecipitation assays showed that both overexpressed and endogenous HBZ protein bound directly to EZH2 (Fig. 2B; Supplementary Fig. S2B). The binding was confirmed even in the presence of nuclease, indicating that the HBZ–EZH2 interaction is independent of the presence of chromatin (Supplementary Fig. S2C). Experiments using deletion mutants of EZH2 (Supplementary Fig. S2D) revealed that HBZ protein binds to the regions of EZH2 where EZH2 binds to other components of PRC2, but not to the catalytic region including the SET domain (Supplementary Fig. S2E). HBZ protein also coimmunoprecipitated with SUZ12 and EED (Supplementary Fig. S2F and S2G). Thus, our findings imply that HBZ protein binds to the PRC2 complex and affects its function.
Figure 2.
HBZ protein alters EZH2 genome-wide distribution and decreases its binding to the TAp73 promoter. A, GSEA plots for mouse CD4+ T cells transduced with WT or mutant HBZ compared with the vector. The normalized enrichment score (NES) and q-value are listed. B, Immunoprecipitation (IP) with anti-Flag antibody (Flag-HBZ) showing interaction between HBZ protein and EZH2 in HEK293T cells. IP was analyzed by SDS-PAGE and immunoblotting (IB). C–E, ChIP-qPCR for EZH2 and H3K27me3 in the TAp73 promoter region. The %Input is shown for WT or HBZ-Tg mouse CD4+ T cells (C), human CD4+ T cells from healthy donors (hCD4) or ATL cell lines (D), and Jurkat cells with stable transduction of WT or mutant HBZ (E; n = 3). F, Histone methyltransferase (HMT) activity on H3K27 among transduced Jurkat cells (n = 3). G and H, Heat maps for genomic regions with enriched ChIP-seq scores for H3K27me3. The score for each region was scaled and clustered based on healthy human donor CD4+ T cells (hCD4; G) or WT mouse CD4+ T cells (H) with transcription start site (TSS) and transcription end site (TES) labeling. In addition to Treg, ATL cells (TL-Om1 and ED; G) and cells from HBZ-Tg mice (H) were analyzed. I, Representative genes found in cluster 2 that were shared between the results from human and mouse cells in G and H, respectively. J, Results of KEGG pathway analysis using the genes in cluster 2 for humans (G). Shared pathways between human and mouse cells are highlighted in red. Statistical values and gene counts calculated by the clusterProfiler are shown. K and L, Scatter plots of mouse cluster 2 genes resulting from combinational analysis of RNA-seq (HBZ-Tg mouse CD4+ T cells relative to WT) and ChIP-seq [relative enrichments in TSS of WT as area under the curve (AUC)] for EZH2 (K) and H3K27me3 (L). Results are plotted as mean ± SD, using Student t test (C) or one-way ANOVA with the post hoc Dunnet test (D–F). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
The epigenetic status of TAp73 promoter regions in ATL cells and in CD4+ T cells of HBZ-Tg mice was evaluated with ChIP quantitative real-time PCR (ChIP-qPCR) and ChIP-seq. Of note, EZH2 recruitment and H3K27me3 were reduced, and H3K27 acetylation (H3K27ac) was increased in the TAp73 promoter in both HBZ-Tg mouse CD4+ T cells and ATL cell lines compared with their corresponding controls (Fig. 2C and D; Supplementary Fig. S2H). This alteration of chromatin state would be expected to result in increased transcription of TAp73, which would explain why TAp73 expression levels are elevated in HBZ-expressing cells (23). It was confirmed that reduced levels of EZH2 and H3K27me3 in the TAp73 promoter were caused by HBZ protein, but not RNA, using HBZ-transduced Jurkat cells (Fig. 2E). We also found that HBZ WT and its mutants did not affect EZH2 enzymic activity (Fig. 2F). All these observations are consistent with the hypothesis that HBZ protein binds to the PRC2 complex but not to the catalytic domain of EZH2.
HBZ Protein Dysregulates Genome-wide Distribution of H3K27me3
Our findings indicate that the HBZ protein changes the transcriptional profiles of some genes by binding to EZH2 without affecting its enzymatic activity. Therefore, we analyzed the genome-wide distribution of EZH2 and H3K27me3 in ATL cells and HBZ-expressing cells. As expected, the accumulation of H3K27me3 at transcription start sites (TSS) changed in a wide variety of genes in both ATL cell lines and CD4+ T cells of HBZ-Tg mice (Fig. 2G and H). In addition, the HBZ-expressing cells exhibited profiles that closely resembled Treg profiles, supporting the previous findings that HBZ protein gives HTLV1-infected cells a Treg-like phenotype (6, 9). Multiple genes were found to be transcriptionally repressed due to accumulation of trimethylation (cluster 1 in Fig. 2G and H), including tumor suppressor genes such as NDRG2, in line with previous reports (14, 24). Concurrently, there were numerous genes in which H3K27me3 at the TSS was decreased, indicating that they were transcriptionally activated (cluster 2 in Fig. 2G and H; Supplementary Tables S1 and S2). Of note, many genes in cluster 2, including TAp73, were shared by both ATL cells and the CD4+ T cells of HBZ-Tg mice (Fig. 2I), and pathway analysis indicated that they are associated with various oncogenic pathways (Fig. 2J; Supplementary Fig. S2I). TAp73 upregulation in HBZ-Tg CD4+ T cells was significantly correlated with reduced levels of EZH2 and H3K27me3 in its promoter (Fig. 2K and L), supporting the idea that EZH2 is a significant regulator of TAp73 expression.
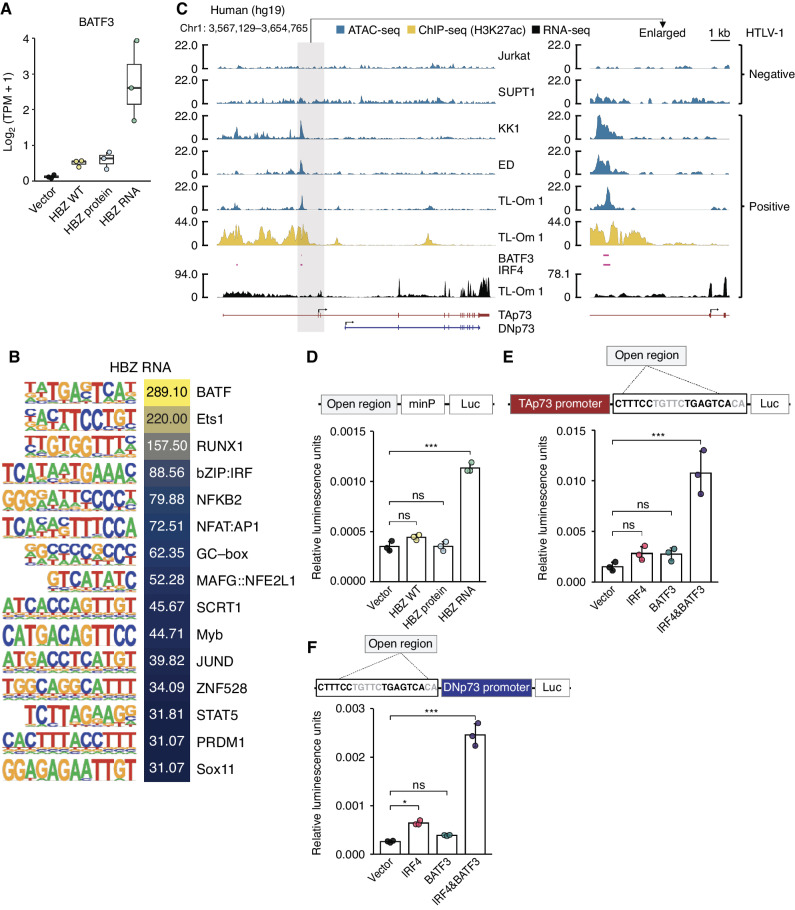
BATF3 Induced by HBZ RNA Activates Both the TAp73 and DNp73 Promoters in Human Cells
Thus far, we have discussed the epigenetic upregulation of TAp73 transcription by HBZ protein via its effects on EZH2. HBZ RNA is thought to upregulate TAp73 by a different mechanism. We found that an open chromatin region was present about 3 kb upstream from the TAp73 TSS, especially in HBZ RNA- and WT-transduced cells (highlighted by gray; Supplementary Fig. S3A). H3K27ac ChIP-seq of HBZ-Tg CD4+ T cells revealed that transcription was activated in this region, where BATF3 and IRF4 can be recruited (Supplementary Fig. S3A, right). Corroborating these findings, BATF3 transcription was drastically upregulated by HBZ RNA, compared with HBZ WT and protein (Fig. 3A) as previously reported (25). Additionally, it was found that the target sequences of BATF are also activated by HBZ RNA (Fig. 3B). We identified a similar region within the TP73 gene body in the human genome (highlighted by gray; Fig. 3C). Recruitment of both BATF3 and IRF4 to this region was confirmed by ChIP-seq and ChIP-qPCR (Fig. 3C; Supplementary Fig. S3B and S3C; ref. 25). BATF3 is known to alter the expression of ATL-associated genes in concert with IRF4 (25, 26). Thus, we next performed promoter reporter assays using the identified region upstream of human TAp73. As expected, HBZ RNA induced this promoter activity (Fig. 3D). The position of the target motifs of BATF3 and IRF4 is closer to both the TAp73 and DNp73 promoters in the human genome (Fig. 3C, left) than in the mouse genome (Supplementary Fig. S3A, left). Intriguingly, the promoters of both human TAp73 and DNp73 were activated by cotransfection of BATF3 and IRF4 (Fig. 3E and F; Supplementary Fig. S3D and S3E), suggesting that HBZ RNA induces DNp73 in human cells, even though it cannot in mouse cells (Supplementary Fig. S1D). Taken together, our data show that HBZ protein and RNA induce TAp73 in different manners, and HBZ RNA activates DNp73 in human cells but not in mouse cells. These findings could collectively explain why both TAp73 and DNp73 were upregulated in patients with ATL (Fig. 1J and K).
Figure 3.
HBZ RNA activates both TAp73 and DNp73 promoters via BATF3-IRF4 transcriptional machinery. A, Transcripts per million (TPM) of BATF3 in HBZ-transduced murine CD4+ T cells (n = 3). B, The top-ranked enriched motifs of HBZ RNA-transduced murine CD4+ T cells with their log2P values from the findMotifsGenome results (HOMER). C, Chromatin accessibility (ATAC-seq), H3K27ac enrichment, BATF3/IRF4 binding regions (ChIP-seq; SRX2548278 and SRX2548284; ref. 25) analyzed by using the ChIP-Atlas (73), and transcripts (RNA-seq) of the TP73 gene in HTLV-1–negative or –positive human T-cell lines. D, Promoter assays of the HTLV-1–specific open region identified in C (hg19 genome region of chr1:3593076–3594185) with a minimal promoter (minP) in HEK293 cells cotransfected with WT or mutant HBZ. The open region was inserted into pNL3.2.CMV after cloning of the genome region as shown in a schematic of construct. E and F, The IRF4/AP-1 motifs identified within the open region (hg19 genome region of chr1:3593076–3594185) were subjected to promoter assays with IRF4 and/or BATF3 induction: for the promoter of TAp73 (E) and the promoter of DNp73 (F; n = 3). A schematic of the assay construct is shown above the corresponding bar plot. Results are plotted with mean ± SD, using one-way ANOVA with the post hoc Dunnet test (D–F). *, P < 0.05; ***, P < 0.001; ns, not significant.
TAp73 Activates EZH2 Transcription
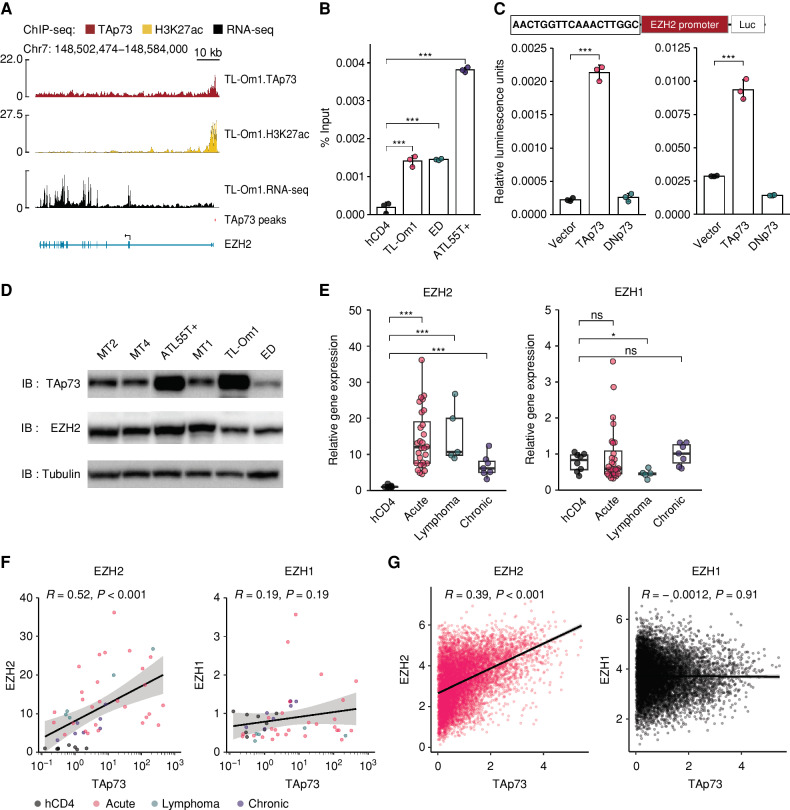
Thus far, we have shown that HBZ RNA appears to increase BATF-activated TAp73 transcription, whereas HBZ protein activates TAp73 by interacting with EZH2. Interestingly, HBZ also appears to cause elevated levels of EZH2 (Supplementary Fig. S3F). High expression of EZH2 with accumulation of H3K27me3 has previously been reported in ATL cells (12–14). HBZ and tax, another HTLV-1 viral oncogene, have been thought to affect EZH2 regulation (27, 28), but the detailed mechanism has been unclear. Because EZH2 was quickly induced by HBZ expression (Supplementary Fig. S3F), we inferred that its transcription might be regulated by HBZ. Indeed, we found that TAp73 was recruited to the promoter region of the EZH2 gene in ATL cell lines (Fig. 4A and B). Luciferase assays revealed that the promoter of TAp73, but no other TP73 isoform, enhances EZH2 promoter activity (Fig. 4C). Indeed, ATL cells endogenously express both TAp73 and EZH2 proteins (Fig. 4D). As previously reported (29), we confirmed the upregulation of the EZH2 gene in ATL patients, while finding that EZH1 expression was not increased (Fig. 4E). Importantly, a positive correlation between TAp73 and EZH2, but not EZH1, was observed in ATL patients (Fig. 4F) and patients with various cancers in TCGA data sets (Fig. 4G), suggesting that TAp73 is associated with the regulation of EZH2 gene expression not only in ATL but also in other types of cancer.
Figure 4.
TAp73 transcriptionally induces EZH2 gene expression. A, TAp73 and H3K27ac enrichment (ChIP-seq) and transcripts (RNA-seq) of the EZH2 gene in TL-Om1 cells. Extracted TAp73 peak is shown as a red bar. B, ChIP-qPCR for TAp73 in the EZH2 promoter region of ATL cell lines relative to healthy human donor CD4+ T cells (hCD4; n = 3). C, Promoter assays using the TP73 motif identified within the TAp73 peak by the ChIP-seq experiment shown in A. Relative luciferase activities with the expression of various TP73 isoforms in HEK293 (left) and Jurkat (right) cells (n = 3). A schematic of the assay construct is shown above the bar plots. D, Immunoblots (IB) of TAp73, EZH2, and Tubulin in HTLV-1–infected cell lines. E, mRNA expression of EZH2 (left) and EZH1 (right) by RT-qPCR in hCD4 (n = 8) and ATL cells (acute type, n = 28; lymphoma type, n = 5; chronic type, n = 7). F and G, Correlation between TAp73 expression and expression of EZH2 (left) or EZH1 (right) in hCD4 and ATL cells (F) and TCGA data (G). Results of Pearson correlation analysis are plotted. Results are plotted as mean ± SD, using one-way ANOVA with the post hoc Dunnet (B and C) or Steel test (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Inactivation of TAp73 Induces Cell Death and Acidification by Lactate Accumulation in ATL Cells
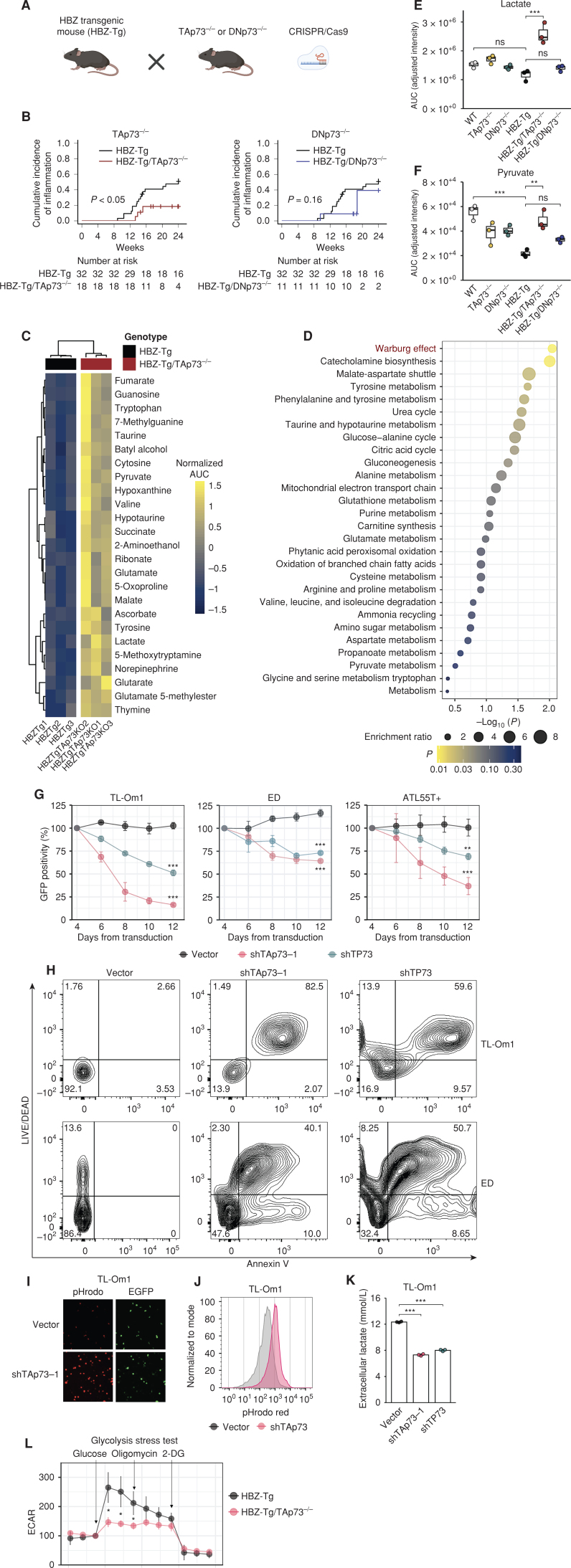
To evaluate the significance of our findings in HBZ-mediated pathogenesis in vivo, we generated two strains of Trp73 knockout mice, TAp73−/− and DNp73−/− (Fig. 5A; Supplementary Fig. S4A), and we crossed them with HBZ-Tg mice. HBZ-Tg mice develop systemic inflammation, such as dermatitis and bronchitis (6). Compared with the frequency of dermatitis in HBZ-Tg mice, the frequency of dermatitis was significantly reduced in HBZ-Tg/TAp73−/− mice, but not in HBZ-Tg/DNp73−/− mice (Fig. 5B). These findings support the interpretation that in mice, the functions of TAp73, not DNp73, are of primary importance in the pathogenicity of HBZ.
Figure 5.
TAp73 inactivation causes ATL cell death and intracellular acidification due to lactate accumulation. A, Mating diagram of HBZ-Tg mice with TP73 knockout mice (TAp73−/− or DNp73−/−) created by CRISPR/Cas9. B, Cumulative incidence of skin inflammation in HBZ-Tg mice (n = 32) compared with HBZ-Tg/TAp73−/− mice (left; n = 18) or HBZ-Tg/DNp73−/− mice (right; n = 11). The P values determined by the Gray test are shown. C and D, Changes in metabolites between HBZ-Tg and HBZ-Tg/TAp73−/− mouse CD4+ T cells. Shown are the top 25 differentially altered metabolites with the normalized area under the curve (AUC; mean-centered and divided by the standard deviation of each variable; C) and enriched metabolite sets (D), both of which were analyzed by using the MetaboAnalyst (78). E and F, Intracellular lactate (E) and pyruvate (F) levels in the murine CD4+ T cells (n = 3). Calculated ion intensities (normalized AUC) are shown. G, GFP competition assay of ATL cell lines with TP73 knockdown (n = 3). Lentiviral vectors for knockdown encoded enhanced GFP (EGFP). The date of lentiviral transduction was counted as day 1. H, Cell viability and apoptosis assay with TP73 knockdown. On day 8 after transduction, ATL cells were analyzed by flow cytometry. Representative dot plots are shown. I and J, Intracellular pH was assessed by pHrodo Red AM in TL-Om1 cells on day 8 after transduction. Fluorescence microscopy photographs with EGFP (indicates lentivirus-transduced cells) and pHrodo Red AM (I) and flow cytometry results (J). K, Extracellular lactate in TL-Om1 cell cultures on day 8 after transduction (n = 3). L, Extracellular acidification rate (ECAR) assessed by metabolic flux assay of CD4+ T cells from HBZ-Tg or HBZ-Tg/TAp73−/− mice (n = 3). Glucose, oligomycin, and 2-deoxyglucose (2-DG) were injected at the indicated time points. Results are plotted as mean ± SD, using one-way ANOVA with post hoc Tukey (E and F), Dunnet test (G and K), or Student t test (L). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Created with BioRender.com.
TAp73 was previously shown to enhance both glycolysis and the pentose phosphate pathway (PPP) by activating transcription of the gene encoding the enzyme that catalyzes each pathway's rate-limiting step (30, 31). Indeed, HBZ-Tg mice showed higher expression levels of G6pdx and Pfkl (Supplementary Fig. S4B). Our HBZ-Tg/TAp73−/− mouse models are useful for assessing the significance of TAp73 in glucose metabolism and its involvement in HBZ-mediated pathogenesis in vivo. Metabolomics analysis showed that CD4+ T cells from HBZ-Tg versus HBZ-Tg/TAp73−/− mice had markedly different metabolite profiles (Fig. 5C). HBZ-Tg/TAp73−/− cells showed similar profiles to WT ones (Supplementary Fig. S4C), suggesting that TAp73 knockout cancels HBZ-driven metabolic alteration. When we analyzed the metabolites that are elevated and accumulated in the cells in HBZ-Tg/TAp73−/− compared with HBZ-Tg mice (Fig. 5C), we found that a metabolite set related to the Warburg effect was highly enriched (Fig. 5D). A more detailed examination of the metabolites associated with the Warburg effect revealed that the intracellular metabolites of glycolysis, such as lactate, pyruvate, and glucose-6-phosphate, were higher in HBZ-Tg/TAp73−/− mice than HBZ-Tg mice (Fig. 5E and F; Supplementary Fig. S4D). This was an unexpected result, because if glycolysis and/or PPP are suppressed by TAp73 knockout, then their end products are expected to be reduced as well.
To examine these findings in more detail, we next performed knockdowns in ATL cell lines using short hairpin RNAs: shTAp73-1 targets only TAp73, whereas shTP73 works against all isoforms of TP73 (Supplementary Fig. S4E–S4G). As expected, we found that EZH2 mRNA was reduced by the knockdown of TAp73 (Supplementary Fig. S4H). We then performed a GFP competition assay in cells transduced with a construct containing shRNA and the GFP gene (Supplementary Fig. S4I), where the proportion of GFP-positive cells changes depending on the effect of knockdown on cell survival (32). Both shRNAs induced necrotic (or late apoptotic) cell death, and intriguingly, this effect was especially robust for shTAp73-1 (Fig. 5G and H). To make sure this was not an off-target effect, we created another TAp73-specific shRNA, shTAp73-2, and obtained similar results (Supplementary Fig. S4J and S4K). Furthermore, intracellular pH became more acidic (Fig. 5I and J; Supplementary Fig. S4L and S4M), and the concentration of extracellular lactate was reduced (Fig. 5K; Supplementary Fig. S4N). We then performed a metabolic flux assay on the glycolytic system. TAp73 knockout in either HBZ-Tg or WT mice led to a markedly reduced extracellular acidification rate (ECAR; Fig. 5L; Supplementary Fig. S4O). Taken together, these observations indicate that inactivation of TAp73 suppresses lactate excretion out of ATL cells, resulting in its intracellular accumulation and cell death.
Lactate Transporters Induced by TAp73 Are Novel Therapeutic Targets for ATL
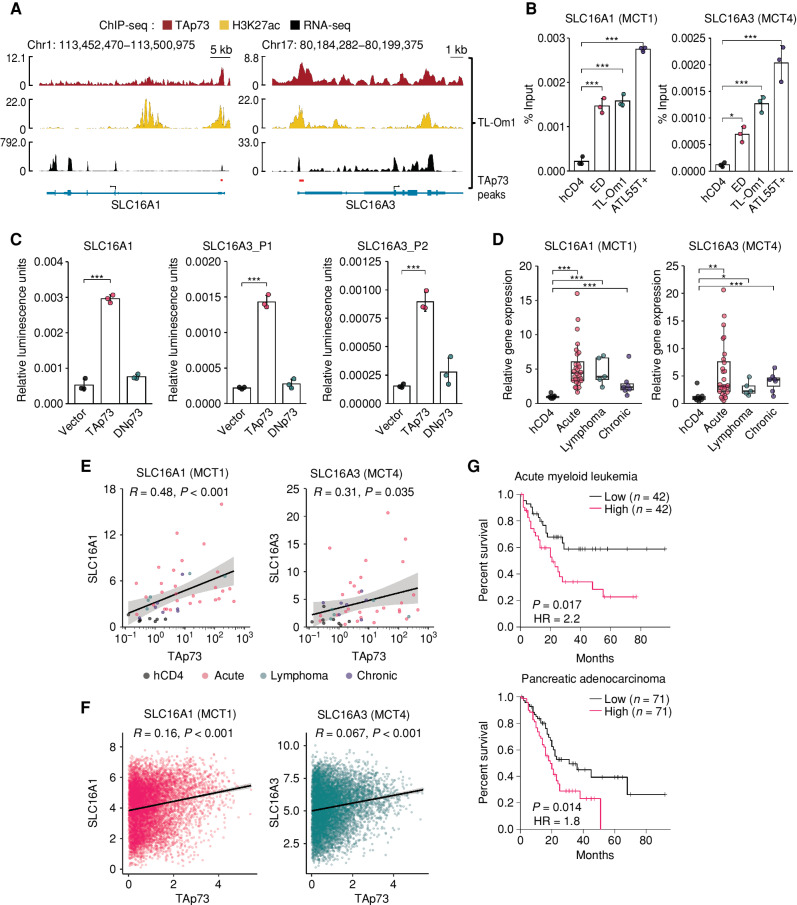
To test the hypothesis that TAp73 regulates lactate transport, we reanalyzed the results of ChIP-seq for TAp73. In ATL cell lines, TAp73 bound to the promoter regions of SLC16A1 and SLC16A3, which encode the lactate transporters MCT1 and MCT4, respectively (Fig. 6A and B). TP73-binding motifs were identified in the extracted ChIP-seq peaks in both genes (red bars in Fig. 6A and sequences in Supplementary Fig. S5A). A promoter assay (Supplementary Fig. S5A) revealed that TAp73, but not other isoforms of TP73, enhanced the promoter activities of both the SLC16A1 and SLC16A3 genes (Fig. 6C; Supplementary Fig. S5B). Corroborating these results, SLC16A1 and SLC16A3 were both upregulated in ATL patients (Fig. 6D), whereas the other MCT family genes, SLC16A7 (encoding MCT2) and SLC16A8 (encoding MCT3), were not (Supplementary Fig. S5C). Furthermore, both SLC16A1 and SLC16A3 expression were positively correlated with TAp73 expression in ATL cells (Fig. 6E).
Figure 6.
TAp73 upregulates both the SLC16A1 and SLC16A3 genes that encode lactate transporters. A, TAp73 and H3K27ac enrichments (ChIP-seq) and transcripts (RNA-seq) of the SLC16A1 (left) and SLC16A3 (right) genes in TL-Om1 cells. Peaks of TAp73 ChIP-seq are shown as red bars in both genes. B, ChIP-qPCR for TAp73 in the SLC16A1 (left) and SLC16A3 (right) promoter regions in ATL cell lines and healthy human donor CD4+ T cells (hCD4; n = 3). C, Promoter assays using the TP73 motif identified within the TAp73 peaks shown in A. Relative luciferase activities for the SLC16A1 promoter (left) and the SLC16A3 promoter (center and right) were examined with TP73 isoform expression in HEK293 cells. (n = 3). Schematics of the assay plasmids are shown in Supplementary Fig. S5A. D, mRNA expression of SLC16A1 (left) and SLC16A3 (right) measured by RT-qPCR in hCD4 cells (n = 8) and ATL patients (acute type, n = 28; lymphoma type, n = 5; chronic type, n = 7). E and F, Correlation between TAp73 expression and expression of SLC16A1 (left) or SLC16A3 (right) in hCD4 cells and ATL patients (E) and TCGA data (F). G, Overall survival of patients with TAp73, SLC16A1, and SLC16A3 high or low gene sets calculated by the GEPIA2 signature scoring (76) on TCGA data (RNA-seq). Group cutoff for splitting each cohort was set as 60% (high) and 40% (low). Statistical values of the log-rank test are shown with hazard ratio (HR). Results are plotted with mean ± SD, using one-way ANOVA with post hoc Dunnet (B and C) or Steel test (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
As expected, TAp73 knockdown in ATL cells resulted in decreased expression of both the SLC16A1 and SLC16A3 genes (Supplementary Fig. S5D and S5E). TCGA data sets also demonstrated that SLC16A1 and SLC16A3 (but not SLC16A7 and SLC16A8) were upregulated in multiple cancer types (Supplementary Fig. S5F–S5I). Positive correlations between the expression levels of TAp73 and SLC16A1 or SLC16A3 were also found in those cancers (Fig. 6F). Notably, acute myeloid leukemia and pancreatic adenocarcinoma patients with high expression of the TAp73/SLC16A1/SLC16A3 three-gene signature had a poorer prognosis with statistical significance (Fig. 6G). To further evaluate the significance of TAp73 in other cancer subtypes, we knocked it down in a chronic myeloid leukemia (CML) cell line using the TAp73-specific shRNA (shTAp73-2 in Supplementary Fig. S4E). The GFP competition assay showed a significant survival reduction in CML (Supplementary Fig. S5J and S5K). These observations imply that TAp73-MCT1/4 metabolic regulation is associated with the prognosis not just of ATL but potentially of various other cancers as well.
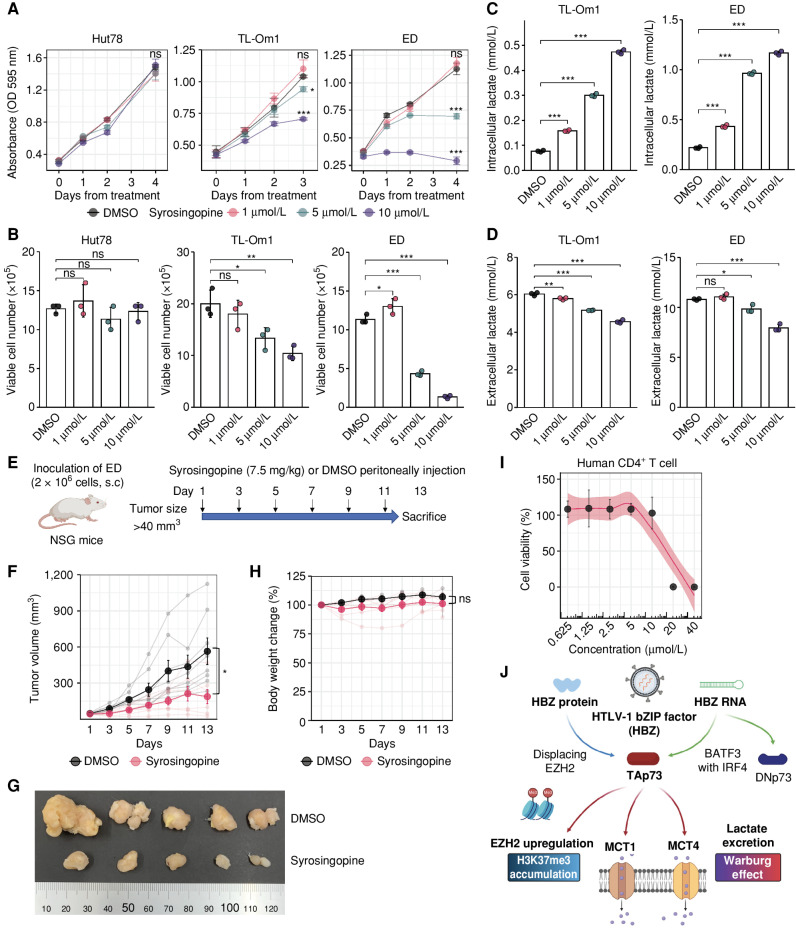
We next evaluated the therapeutic efficacy of syrosingopine, which is an MCT1/4 dual inhibitor (33, 34) on ATL cells, and found that it induced cell death in a dose-dependent manner (Fig. 7A and B; Supplementary Fig. S6A–S6C). Importantly, syrosingopine treatments led to intracellular accumulation of lactate (Fig. 7C; Supplementary Fig. S6D) and reduced the concentration of extracellular lactate (Fig. 7D; Supplementary Fig. S6E) in ATL cells. Syrosingopine significantly suppressed the growth of ATL cells transplanted into immunodeficient NOD/SCID/IL2Rgnull (NSG) mice without body weight loss (Fig. 7E–H). We further evaluated the safety profile of syrosingopine in human CD4+ T cells from healthy donors. Cell toxicity was not observed at 5 μmol/L (Fig. 7I), whereas the same concentration of the drug significantly suppressed the growth of ATL cell lines (Fig. 7A and B; Supplementary Fig. S6A–S6C), suggesting that syrosingopine has leukemic cell-specific toxicity. Taken together, these results show that MCT1/4 dual inhibition yields antitumor effects by blocking lactate excretion from ATL cells in vitro and in vivo.
Figure 7.
Efficacy of the MCT1/4 inhibitor syrosingopine on ATL cells. A and B, Cell proliferation assay (A) and viable cell numbers (B) for HTLV-1–unifected T cells (Hut78) or ATL cells (TL-Om1 and ED) treated with syrosingopine (1 μmol/L, 5 μmol/L and 10 μmol/L; n = 3). Values in comparison with the dimethyl sulfoxide (DMSO) group are shown. C and D, ATL cells expel less lactate when treated with syrosingopine. Lactate was examined intracellularly (C) or extracellularly (D) after 4 days of syrosingopine treatment. E–H, Therapeutic efficacy of syrosingopine in NOD/SCID/IL2Rgnull (NSG) mice after inoculation with ATL cells (ED). Experimental outline for the in vivo experiment to compare syrosingopine with DMSO (E). Tumor volumes were measured every 2 days in NSG mice treated with syrosingopine (n = 7) or DMSO (n = 8; F). Representative tumors resected from the mice are shown in G. Body weight changes of NSG mice treated with syrosingopine (n = 7) or DMSO (n = 4; H). I, Viability of human CD4+ T cells from healthy donors undergoing syrosingopine treatment (n = 3). J, Graphical summary of this study. HBZ protein and RNA both induce the Warburg effect and H3K27me3 accumulation by activating TAp73 expression. Results are plotted as mean ± SD, using one-way ANOVA with post hoc Dunnet test (A–D) or Student t test (F and H). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Created with BioRender.com.
DISCUSSION
In many cancer tissues, the metabolism of glucose is often dysregulated in order to fuel cell proliferation. Activation of several protooncogenes and mutations of certain tumor suppressor genes have been shown to reprogram the glycolysis process (35, 36). Myc increases the expression of many metabolic enzymes, including LDH-A, and several enzymes required for nucleotide biosynthesis (36). It has also been reported that constitutive activation of Akt increases the surface expression of glucose transporters and stimulates the expression and enzyme activity of the factors associated with glycolysis (35). Multiple lines of evidence indicate that ATL shares multiple hallmark capabilities with other cancers (37); however, there are few studies reporting the involvement of altered glucose metabolism in the oncogenic activity of HTLV-1.
In this study, we show that an isoform of the TP73 gene containing the transactivation domain, TAp73, is induced by HBZ and plays critical roles in the excretion of lactate (Fig. 7J), which is a waste metabolite of glycolysis (38). Several studies previously showed that TAp73 transactivates genes associated with glycolysis, such as GP6D and PFKL, suggesting that TAp73 might have a role in the Warburg effect (31). Expression of these genes was also upregulated in HBZ-transgenic cells (Supplementary Fig. S4B), and more notably, the expression of the lactate transporters MCT1 and MCT4 was significantly upregulated, suggesting that TAp73 both accelerates the glycolytic cascade and boosts the efflux of metabolites out of the cells. Interestingly, the positive correlation between TAp73 and SLC16A1/3 expression is observed in other cancer types, and this signature is related to their prognosis with statistical significance (Fig. 6F and G). These observations imply that the promotion of lactate excretion by TAp73-induced MCT1/4 is a common feature of many cancers.
We also found that in addition to inducing glycolysis and lactate transport, TAp73 induces EZH2 transcription (Figs. 4A–C and 7J). Previous studies have shown that EZH2 is highly expressed in ATL cells and alters their epigenetic landscape to repress the expression of several tumor suppressor genes such as NDRG2 and ZEB1 (14). Because inhibitors of EZH2 have been proven to be effective against ATL cells (29), EZH2 is recognized as one of the key molecules involved in ATL leukemogenesis. Our results suggest that transcriptional upregulation of TAp73 by HBZ is an important mechanism for inducing constitutive expression of EZH2 in ATL cells and even in HTLV-1–infected cells. Surprisingly, we also identified a strong correlation between TAp73 and EZH2 transcription levels in the TCGA data set (Fig. 4G), suggesting that TAp73 is a common transactivator of EZH2 in many types of cancer, although concluding this would require further analysis of TAp73 and EZH2 proteins in a larger number of cases. Thus, TAp73 is associated with “nonmutational epigenetic reprogramming” described in the recently revised hallmarks of cancer (2).
Clonal expansion of infected cells is crucial for the persistent infection of HTLV-1 in vivo and eventually leads to malignant transformation of infected cells. HBZ plays important roles in the proliferation and survival not only of ATL cells but also of infected nonleukemic cells. We and others have reported the distinct functions of HBZ protein and HBZ RNA (5, 10, 39). RNAs imbued with both coding and noncoding functions are known to exist in bacteria, plants, and animals and are referred to as “bifunctional RNAs” or “coding and noncoding RNAs (cncRNAs)” (40). In mammals, several mRNAs have been proven to harbor noncoding functions independent of the proteins they encode. For example, Steroid receptor activator (SRA), a functional RNA, and SRAP, a translated protein, counteract each other in myogenic differentiation (41). A well-known tumor suppressor gene, TP53, also has coding and noncoding functions; p53 mRNA inactivates the E3 ubiquitin ligase MDM2, a negative regulator of p53 protein, and consequently controls p53 responses under genotoxic stresses (42). Each case of bifunctional RNA is thought to be a native machinery for fine-tuning its own activities. In contrast, HBZ RNA and protein regulate the function and/or expression of numerous host factors and promote the clonal proliferation of HTLV-1–infected cells. Here, we show that both HBZ protein and HBZ RNA strongly induce TAp73 expression through different mechanisms: HBZ protein diminishes recruitment of EZH2 to the TAp73 promoter, whereas HBZ RNA upregulates BATF3, which activates the TAp73 promoter (Fig. 7J). Thus, HTLV-1 has redundant mechanisms to induce TAp73. HBZ RNA was found to change its subcellular localization depending on its promoter activity (15), and it is important for HBZ RNA to localize in the nucleus to promote cell growth. Considering that translation into protein is generally executed in the cytoplasm (43), it is possible that HBZ RNA and HBZ protein are differentially expressed depending on the activity of the HBZ promoter. Redundant mechanisms for TAp73 induction by HBZ RNA and protein might be useful to maintain TAp73 expression regardless of cell status. Recently, it has been reported that an intragenic super-enhancer in the TP73 gene is active in ATL cells, and it contributes to the high expression of TP73 and cell growth (44). Because HBZ protein reduces the level of H3K27me3 in the same genomic region in human T cells (Fig. 2E), it is suggested that modulation of the epigenetic status by HBZ is involved in the formation of the TP73 intragenic super-enhancer, which supports its continuous expression.
The role of TP73 in oncogenesis has been mainly associated with the expression of DNp73, a dominant-negative isoform with antiapoptotic properties (17). Our expression analysis revealed increased expression of both TAp73 and DNp73 in ATL cells (Fig. 1J and K), but we focused on the unexpected result that knockdown of TAp73 alone reduced proliferation of the ATL cell lines we used (Fig. 5G; Supplementary Fig. S4K). Previous genomic studies reported that ∼20% of ATL cases lack the transactivation domain of the TP73 gene due to structural aberration (45), suggesting that in those cases, the antiapoptotic effect of DNp73 is more important for the survival of ATL cells than the transactivating properties of TAp73.
Our results suggest that BATF3, induced by HBZ RNA, is associated with the activation of its target motif near to the human TP73 gene (Fig. 3C–F). Similar AP-1-IRF composite element (AICE) motifs are absent near the mouse Trp73 gene. This difference is a possible reason for our findings that DNp73 is induced by HBZ RNA in human T cells (Fig. 1K) but not in mouse T cells (Supplementary Fig. S1D and S1E). The fact that HBZ RNA does not induce DNp73 expression in mice may explain why TAp73, not DNp73, was important in the development of dermatitis (Fig. 5B). In humans, the story may be more complicated: we have shown that TAp73 plays an important role in ATL, yet DNp73 was also reported to function as an oncogenic protein via its inhibitory effect on TAp73- or TP53-mediated apoptosis (46), and we found that DNp73, like TAp73, responds to the induction of BATF3 by HBZ RNA. At this moment, the molecular mechanisms for induction of BATF3 by HBZ RNA remain unknown, and they will need to be elucidated in the future.
In summary, TAp73, induced by HBZ, contributes to both the Warburg effect and epigenetic reprogramming in HTLV-1–infected cells via induction of MCT1/4 and EZH2, respectively. These signatures are commonly observed in multiple types of cancer, implying that these molecules are promising targets for wide-spectrum anticancer strategies.
METHODS
Oligonucleotides are listed in Supplementary Table S3, and experimental reagents and resources with identifiers are summarized in Supplementary Table S4.
Patients
Peripheral blood mononuclear cells were isolated from healthy donors, acute- and chronic-type ATL patients with Ficoll-Paque Plus (GE Healthcare). For lymphoma-type ATL, we obtained primary ATL cells from the enlarged lymph nodes. Isolation of CD4+ T cells was performed using the Human CD4 T Lymphocyte Enrichment Set-DM (BD Biosciences). All samples were obtained with informed consent for research use. This study was conducted in accordance with the principles in the Declaration of Helsinki. We obtained written informed consent from ATL patients and healthy donors with approval by the Institutional Ethics Committee of Kumamoto University (approval number Genome 297) and Kyoto University (approval number G204).
Cell Lines
HTLV-1–negative human T-cell lines [Jurkat (male) and Hut78 (male)] and ATL cell lines [TL-Om1 (male), ED (male), ATL55T+ (unspecified), MT1 (male), and HPB-ATL-2 (female)] were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). ATL55T+ was incubated with recombinant human interleukin 2 (100 U/mL; PeproTech) because of its dependency. For HEK293 and HEK293T cells (unspecified), DMEM was used with 10% FBS. Stably HBZ-transduced lines were generated from Jurkat cells with G418 selection (1 mg/mL; Nacalai Tesque), in which the coding sequence of HBZ or HBZ mutants was subcloned into pME18Sneo vector and transduced by using Neon Transfection System Kit (Thermo Fisher Scientific) as described previously (7, 47). All cell lines were incubated at 37°C in a 5% CO2 humidified incubator and routinely tested for Mycoplasma spp. The source of each cell line and authentication are listed in Supplementary Table S4.
Mice
C57BL/6J (CLEA Japan) and NOD/SCID/IL2Rgnull (NSG; The Jackson Laboratory) mice were purchased. The HBZ-Tg were developed from the C57BL/6J mice and genotyped as previously described (5). At sacrifice, primary splenocytes were collected, and CD4+ T cells were then isolated with the Mouse CD4 T Lymphocyte Enrichment Set-DM (BD Biosciences). All animal procedures in this study were conducted in accordance with the Institutional Animal Care and Use Committee at Kumamoto University (approval number A2019-056 and A2021-135) and Kyoto University (approval number G204).
Generation of TAp73−/− and DNp73−/− Mice
The CRISPR/Cas9 system was utilized for isoform-specific knockout of the Trp73 gene in C57BL/6J mice. Two types of small RNAs, target-recognizing CRISPR RNA (crRNA) designed with CHOPCHOP (48), and auxiliary transactivating crRNA (tracrRNA), were purchased from FASMAC as listed in Supplementary Table S3. We delivered TrueCut Cas9 Protein v2 (Thermo Fisher Scientific) together with the crRNA and tracrRNA into in vitro–fertilized eggs of C57BL/6J mice according to the institutional procedure of Kyoto University (49). Both TAp73 and DNp73 knockout genotypes were obtained with 1bp insertions on the crRNA recognizing region, which results in truncating (frameshift) mutations (Supplementary Fig. S4A). Genotyping for each knockout isoform was performed with restriction enzyme digestion of PCR-amplified DNA. For TAp73, genomic DNA was amplified with TAp73-genotype-F and TAp73-genotype-R primers (Supplementary Table S3), followed by BstBI (New England Biolabs) digestion, whereas DNp73-genotype-F and DNp73-genotype-R primers (Supplementary Table S3), and MseI (New England Biolabs) were similarly utilized for DNp73 genotyping.
Murine Xenograft Model
2 × 106 ED cells were subcutaneously inoculated in the right lower back of 6-week-old female NSG mice. Tumor growth was followed until the tumor reached a size of 40 mm3, at which point (day 1), the NSG mice were randomly assigned to either the syrosingopine (7.5 mg/kg; MedChemExpress) or the dimethyl sulfoxide (DMSO) group. Treatments were injected intraperitoneally every other day for a total of six doses. Tumor sizes were also measured with calipers every other day until day 13. The xenograft model study was conducted under the condition that mice be euthanized if the tumor size exceeded 2,000 mm3. At the end of the observation, the tumors were also weighed.
Retroviral Transduction of Murine CD4+ T Cells
Retroviral vectors containing the GFP and HBZ WT or HBZ mutant were transduced into murine CD4+ T cells as described previously (5, 10). GFP-positive cells were collected for subsequent deep-sequencing.
RNA-seq
Biological triplicates were adopted for RNA-seq in this study. Total RNA was collected with the ReliaPrep RNA Miniprep System (Promega). Libraries were generated with the TruSeq stranded mRNA Library Prep kit (Illumina). Sequencing was performed on the Illumina NovaSeq 6000 with a standard 100-bp paired-end read protocol.
Analysis of RNA-seq
In addition to the sequencing data in this study, RNA-seq data were obtained from the following public archives: DRP006148 (50) for CD4+ T cells of HBZ-Tg and WT mice; SRP301994 (15) for TL-Om1 cells; GSE74246 (51) and GSE162712 (52) for normal human CD4+ T cells (hCD4); and SRP042199 (53), GSE143986 (54), and DRP006659 (55) for ATL cells. After fastq files were cleansed with Trim Galore (https://github.com/FelixKrueger/TrimGalore), obtained reads were mapped to the hg19 or mm10 genome sequence. To align them, HISAT2 (56) was used with the following options: –dta-cufflinks, –no-discordant, –no-mixed and –rna-strandness FR. The aligned data were then sorted by samtools (57). After counting reads in features with htseq-count of HTSeq (58), differentially expressed genes were identified if the fold change was greater than 1.1 and the adjusted P value (Padj) was less than 0.05 in the DESeq2 analysis (59). The results were then subjected to GSEA (60) using clusterProfiler (61). Transcripts per million (TPM) were calculated using TPMCalculator (62).
qRT-PCR
Total RNA was purified using the ReliaPrep RNA Miniprep System (Promega) or TRIzol Reagent (Thermo Fisher Scientific) and then converted to cDNA with SuperScript IV (Thermo Fisher Scientific). Gene expression was assessed using FastStart Universal SYBR Green Master (Roche) on the StepOnePlus Real-Time PCR System (Applied Biosystems). Primers for qRT-PCR are listed in Supplementary Table S3.
Immunoprecipitation and Immunoblotting
For immunoprecipitation, antibodies against Flag (Sigma-Aldrich, #F3165), HA (MBL International, #M180-3), and HBZ [rabbit polyclonal antibody generated by immunizing with HBZ-peptides (CRGPPGEKAPPRGETH and QERRERKWRQGAEKC); MBL International; ref. 50], along with normal rabbit IgG, were used. Immunoblotting was conducted with antibodies against HBZ described above, Flag (Sigma-Aldrich, #F7425), HA (MBL International, #561-5), α-Tubulin (Sigma-Aldrich, #T6199), EZH2 (Cell Signaling Technology, #5246S), TAp73 (Abcam, #ab14430), and DNp73 (Novus Biologicals, #NBP2-24873). For DNase treatment, ATL55T+ cell lysates were incubated with or without 2 μL DNase I Solution (2,500 U/mL; Thermo Fisher Scientific) for 30 minutes at 37°C.
ChIP
ChIP assays were conducted with the use of the SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology) according to the manufacturer's instructions. Briefly, cross-linked chromatin prepared from 4 × 106 cells was immunoprecipitated overnight with antibodies against EZH2 (Cell Signaling Technology, #5246S), H3K27me3 (Cell Signaling Technology, #9733S), IRF4 (Cell Signaling Technology, #4964S), BATF3 (R&D Systems, #AF7437), or TAp73 (Abcam, #ab14430). The immunocomplexes were then conjugated with ChIP-Grade Protein G Magnetic Beads (Cell Signaling Technology) for 2 hours at 4°C. After 4 washes, the bead-bound DNA was eluted by gentle vortexing (1,200 rpm) for 30 minutes at 65°C. Purified DNA was quantified by ChIP-qPCR in comparison with a 2% input control. The primers for ChIP-qPCR are listed in Supplementary Table S3. The eluted DNAs and the input were also subjected to ChIP-seq. The TruSeq ChIP Library Preparation Kit (Illumina) was used for library preparation. Libraries were sequenced on the Illumina NovaSeq6000 platform with a standard 150-bp paired-end read protocol.
ATAC-seq
Cells were incubated with Nextera Tn5 Transposase and 2× TD reaction buffer of the Illumina Tagment DNA Enzyme and Buffer Small Kit (Illumina) for 30 minutes at 37°C, according to our previous report (55). Sequencing was performed on the Illumina HiSeq 4000 with a standard 100-bp paired-end read protocol.
Cleansing and Mapping of DNA Sequencing Data (ATAC-seq and ChIP-seq)
As with the RNA-seq analysis, fastq files of both ATAC-seq and ChIP-seq data were trimmed and qualified by the Trim Galore. Mapping to the corresponding genome was carried out by Bowtie2 with the “–very-sensitive” option (63). To remove mitochondrial reads and reads that aligned twice or more, we removed “MT” and/or “XS” tagged reads using samtools. Subsequently, reads with a mapping quality of more than 30 were selected using the samtools view with the “-q 30” option. BAM files were also cleaned up by removing PCR duplicates with Picard (MarkDuplicates, http://broadinstitute.github.io/picard/) and the ENCODE Blacklist genome regions (64) using BEDtools (intersectBed; ref. 65).
Peak Calling of ATAC-seq Data in Retrovirally HBZ-Transduced Mice Samples
MACS2 (66) was used for peak calling with the following options: “–nomodel –nolambda –keep-dup all –call-summits.” We further used HOMER (67) for subsequent analysis including motif analysis. Peak bed files from the MACS2 results were converted to a HOMER bed file using the bed2pos.pl tool. After merging the peaks with mergePeaks, we counted each extracted peak using HTSeq. Using these counts with DESeq2, we compared samples from cells transduced with WT HBZ, mutant HBZ, or vector alone. Statistically significant peaks (defined as peaks with Padj less than 0.05) were subjected to motif analysis using findMotifsGenome.pl of HOMER with the “-size 200 -mask” options.
Peak Calling of ChIP-seq Data
Using the corresponding input-sequenced file as a control, ChIP-seq peaks were assessed by MACS2 (66) with the “-p 1e-3” option. Extracted peaks were then annotated to the nearest genes of the hg19 by using the Genomic Regions Enrichment of Annotations Tool (GREAT; ref. 68). We also utilized publicly available ChIP-seq databases with the following ID: CD4+ T cells of HBZ-Tg and WT mice for H3K27ac (DRX028570) with input (DRX028565; ref. 8), hCD4 for H3K27ac (SRX1142198) with input (SRX1142248; ref. 69); TL-Om1 cells for H3K27ac (SRX2026362) with input (SRX2026363; ref. 70), human Tregs for H3K27me3 (SRX088861) with input (SRX088829; ref. 71), murine Tregs for H3K27me3 (SRX1241151) with input (SRX1241155; ref. 72), and the ChIP-Atlas (73) peak-call bed files of murine CD4+ Th2 cells for IRF4 and BATF3 (SRX2646182 and SRX1998451; ref. 74) and KK1 cells for IRF4 and BATF3 (SRX2548278 and SRX2548284; ref. 25). For generating the heat maps of H3K27me3 genomic distribution, we used deepTools (75). First, we extracted peaks of each H3K27me3 ChIP-seq sample compared with the corresponding input sample by using the bamCompare tool with the “–operation subtract” option. To calculate enriched scores per genome region for each transcript, the bigWig files were then analyzed using the computeMatrix tool (reference-point mode) with the “–beforeRegionStartLength 3000 –regionBodyLength 5000 –afterRegionStartLength 3000 –skipZeros” options. Subsequently, heat maps were generated using the plotHeatmap tool with the “–kmeans 5 –sortRegions=descend” options. The clustering was carried out based on human healthy donor or WT mouse CD4+ T cells. The cluster 2 genes (Supplementary Tables S1 and S2) were subjected to KEGG pathway analysis by using the clusterProfiler. For RNA-seq and ChIP-seq combined scatter plots, we first calculated the area under the curve (AUC) of cluster 2 (4,297 genes) by using the matrix file extracted from the plotHeatmap tool above. Then, the AUC values were compared between WT and HBZ-Tg mice and combined with fold changes calculated from RNA-seq data.
Analysis of TCGA Data
Raw data were obtained from the official website of TCGA Pan-Cancer (PANCAN). We analyzed the following data sets: (i) tcga_RSEM_gene_tpm, (ii) tcga_rsem_isoform_tpm, and (iii) TCGA_survival_data_2.txt. We also utilized a web server called GEPIA2 to analyze and visualize TCGA data (76). Overall survival was analyzed with the log-rank test and signature-scoring method of GEPIA2. To split the patient cohort of TCGA data, the group cutoff was set as 60% (high) and 40% (low) by using TAp73, SLC16A1, and SLC16A3 signatures scores. The abbreviations used in TCGA are found at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations.
Histone Methyltransferase Activity Assay
Using the EpiQuik Nuclear Extraction Kit (EpigenTek), we isolated nuclear proteins from Jurkat cells that were stably transduced with WT or mutant HBZ. The nuclear extracts were then subjected to assays of histone methyltransferase activity that specifically target histone H3 at lysine 27. We used the EpiQuik Histone Methyltransferase Activity/Inhibition Assay Kit (H3-K27; EpigenTek). We measured the absorbance at 450 nm for each sample and generated a standard curve. The histone methyltransferase activity (OD/h/mg) was calculated according to the manufacturer's instructions.
Luciferase Assay
The vectors for the luciferase assay, pGL4.10[luc2] and pNL3.2.CMV, were purchased from Promega. The latter was edited for each promoter assay. For Fig. 3D, the hg19 genome region of chr1:3593076–3594185 was cloned using the Zero Blunt Cloning Kit (Thermo Fisher Scientific) and inserted between the KpnI and XhoI sites of pNL3.2.CMV. For all remaining luciferase assays in this study, the minimal promoter (minP) was removed using the In-Fusion Snap Assembly Master Mix (TaKaRa), and the appropriate sequences (enumerated in Supplementary Table S3) were inserted between the KpnI and XhoI sites of pNL3.2.CMV. Ectopic gene expression in the luciferase study was induced by pME18S or pCAGGS containing the corresponding coding sequence. HEK293 or Jurkat cells were transfected with the indicated plasmids using TransIT (for HEK293) or the Neon transfection system (for Jurkat). After 24 hours of incubation, the transfected cells were subjected to luciferase assay.
Lentiviral Transduction of ATL Cell Lines
For the generation of lentiviral particles, we transduced psPAX2, pCMV-VSV-G, and shRNA-encoding lentiviral vectors (Sigma-Aldrich) into HEK293T cells with Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). For subsequent GFP competition assays, the MISSION TRC2 pLKO.5-puro plasmid (Sigma-Aldrich) was modified to replace the puromycin-resistance gene with the EGFP gene using the In-Fusion Snap Assembly Master Mix (TaKaRa). Forty-eight hours after transfection, the supernatant was collected and ultracentrifuged (25,000 rpm) for 2 hours at 4°C. Enriched lentiviruses were infected into ATL cell lines using RetroNectin (TaKaRa) with spin infection (3,000 rpm) for 1 hour.
Flow Cytometry and GFP Competition Assay
After lentiviral transduction (counted as day 1), the GFP positivity of ATL cell lines was analyzed by flow cytometer (FACSVerse) every other day from days 4 to 12. GFP was evaluated only in live cells. The GFP-positive rate on day 4 was used as a baseline to evaluate changes in the positivity rate. The cells were analyzed using the LIVE/DEAD Fixable Dead Cell Stain Kit (Thermo Fisher Scientific), Alexa Fluor 647 Annexin V (BioLegend), and pHrodo Red AM Intracellular pH Indicator (Thermo Fisher Scientific). Extracted data were analyzed by the FlowJo software (BD Bioscience).
Lactate Assay
The Lactate Assay Kit-WST (Dojindo) was used according to the manufacturer's instructions. Intracellular lactate was assayed after the lysis of cells with 0.1% Triton X-100 (Nacalai Tesque). After incubation at 37°C for 30 minutes, the absorbance at 450 nm was measured by using the TriStar LB941 and MikroWin 2000 version 4.41 (Berthold Technologies). In experiments in which ATL cell lines were treated with syrosingopine, extracellular and intracellular lactate were measured after treatment with syrosingopine for 4 days, respectively.
Metabolomic Analysis by Gas Chromatography–Mass Spectrometry
Isolated murine CD4+ T cells were subjected to gas chromatography–mass spectrometry (GC-MS) as described previously (77). Briefly, sonicated samples with 100% methanol were evaporated using a centrifugal evaporator (DNA120OP230, Thermo Fisher Scientific) and lyophilized (FD-1-84A, FTS Systems). Then, the samples were derivatized with methoxamine/pyridine and N-methyl-N-(trimethylsilyl)trifluoroacetamide and analyzed the GCMS-TQ8050 (Shimadzu). The chromatograms and mass spectra were evaluated with the GCMSsolution software 4.50 (Shimadzu). Compounds were determined with the Smart Metabolites Database Ver.2 (Shimadzu). The data obtained were then scaled (mean-centered and divided by the standard deviation of each variable) and analyzed using MetaboAnalyst (78).
Metabolic Flux Assay
After coating Seahorse XFe24 Cell Culture Microplates (Agilent) with Poly-D-lysine hydrobromide (FUJIFILM Wako Pure Chemical), we seeded isolated and viable mouse CD4+ T cells at 1 × 106 cells per well. The cells were incubated in Seahorse XF RPMI medium, pH 7.4 (Agilent) supplemented with 2 mmol/L Seahorse XF Glutamine Solution (Agilent) at 37°C. We used the Seahorse XF Glycolysis Stress Test Kit (Agilent) according to the manufacturer's instructions. The assay was carried out on the Seahorse XFe24 Analyzer (Agilent). Extracted data were analyzed using Seahorse Wave Desktop Software (Agilent).
Cell Proliferation and Viability Assay
Cell lines were treated with syrosingopine (MedChemExpress, an MCT1/4 inhibitor). At first, syrosingopine was dissolved in DMSO. The dissolved syrosingopine or DMSO alone was finally diluted 100-fold in RPMI medium with cell lines and used for further experiments. Cells were then seeded into a 96-well plate without removing syrosingopine and DMSO throughout the experiments. To assess cell growth, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was utilized as previously reported (10). The MTT assay was started 3 hours after the seeding (counted as day 0). Cell viability was assessed by the Countess Automated Cell Counter (Thermo Fisher Scientific) on day 4.
Statistical Analysis and Data Visualization
The Student t test (two-sided) was used for comparisons between the two groups, and differences were defined as statistically significant for P < 0.05. For intercomparison of more than 2 groups, a one-way ANOVA followed by a post hoc test [Tukey, Dunnet (parametric) or Steel (nonparametric)] was applied. To assess the association between targeted gene expressions, we used Pearson correlation analysis. In mouse experiments, the sample size was not preselected and no inclusion nor exclusion criteria were used. The cumulative incidence of inflammation was analyzed by the Gray test. The probability of survival was estimated by the Kaplan–Meier method, and hazard ratios (HR) were estimated with the Cox regression model. All analyses were conducted using R Statistical Software (v4.1.2). The bar and box plots contain the mean value and standard deviation calculated from at least three biological replicates. To generate these plots, we used ggplot2 and pheatmap. To visualize deep-sequencing data, we used deepTools (75) and SparK (https://github.com/harbourlab/SparK). Schematic diagrams in this study were created with BioRender.com.
Data Availability
Sequencing data in this study have been deposited with links to BioProject accession number PRJDB15617 in the DDBJ BioProject database.
Supplementary Material
Human cluster 2 genes whose H3K27me3 in TSS's was decreased.
Mouse cluster 2 genes whose H3K27me3 in TSS's was decreased.
Oligonucleotides used in this study.
Experimental reagents and resources with identifiers used in this study.
Transcriptional regulation by HBZ protein and RNA and the status of TP73 expression.
HBZ protein alters epigenetic regulation of TAp73 by EZH2.
Regulation of TP73 by BATF3 and IRF4.
TAp73 significance in HBZ pathogenesis and its effect on cellular metabolism.
Transcriptional regulation of SLC16A1 and SLC16A3 by TAp73, and mRNA expression levels of MCT-coding genes among various cancers.
Efficacy of the MCT1/4 inhibitor syrosingopine on ATL cells.
Acknowledgments
We thank Dr. Linda Kingsbury for proofreading this manuscript. We also thank Chiho Onishi, Miho Matsumoto, Saiko Tokunaga, Chiaki Ohama, and Ryu Yamashita for their technical assistance with the experiments. This work was supported by a grant from the Project for Cancer Research and Therapeutic Evolution (P-CREATE; 20cm0106306h0005 to M. Matsuoka; 19cm0106611h0003 to J. Yasunaga), the Research Program on Emerging and Re-emerging Infectious Diseases (20fk0108088h0002 to M. Matsuoka) from the Japan Agency for Medical Research and Development (AMED); Science and Technology Platform Program for Advanced Biological Medicine (21am0401003h0003 to J. Yasunaga) from AMED, JSPS KAKENHI (19H03689 to M. Matsuoka; 20H03514 to J. Yasunaga), and the Friends of Leukemia Research Fund (to K. Toyoda.). This study was also supported in part by the JSPS Core-to-Core Program A, Advanced Research Networks.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
K. Toyoda reports personal fees from Friends of Leukemia Research Fund during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
K. Toyoda: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Yasunaga: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing. T. Shichijo: Data curation, formal analysis, investigation, methodology. Y. Arima: Resources, software, methodology. K. Tsujita: Resources. A. Tanaka: Resources, data curation, software, formal analysis, methodology. T. Salah: Data curation, formal analysis. W. Zhang: Data curation, formal analysis. O. Hussein: Data curation, formal analysis. M. Sonoda: Data curation, formal analysis. M. Watanabe: Data curation, formal analysis. D. Kurita: Data curation, formal analysis. K. Nakashima: Resources, data curation, formal analysis. K. Yamada: Resources, data curation, formal analysis. H. Miyoshi: Resources, data curation, formal analysis, supervision. K. Ohshima: Resources, data curation, formal analysis, supervision. M. Matsuoka: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 3. Warburg O. On the origin of cancer cells. Science 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 4. Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11:325–37. [DOI] [PubMed] [Google Scholar]
- 5. Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Nat Acad Sci U S A 2006;103:720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog 2011;7:e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kinosada H, Yasunaga JI, Shimura K, Miyazato P, Onishi C, Iyoda T, et al. HTLV-1 bZIP factor enhances T-cell proliferation by impeding the suppressive signaling of co-inhibitory receptors. PLoS Pathog 2017;13:e1006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yasuma K, Yasunaga J, Takemoto K, Sugata K, Mitobe Y, Takenouchi N, et al. HTLV-1 bZIP factor impairs anti-viral immunity by inducing co-inhibitory molecule, T cell immunoglobulin and ITIM domain (TIGIT). PLoS Pathog 2016;12:e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao T, Satou Y, Sugata K, Miyazato P, Green PL, Imamura T, et al. HTLV-1 bZIP factor enhances TGF-β signaling through p300 coactivator. Blood 2011;118:1865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitobe Y, Yasunaga J, Furuta R, Matsuoka M. HTLV-1 bZIP factor RNA and protein impart distinct functions on T-cell proliferation and survival. Cancer Res 2015;75:4143–52. [DOI] [PubMed] [Google Scholar]
- 11. Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell stem cell 2014;14:735–51. [DOI] [PubMed] [Google Scholar]
- 12. Sasaki D, Imaizumi Y, Hasegawa H, Osaka A, Tsukasaki K, Choi YL, et al. Overexpression of enhancer of zeste homolog 2 with trimethylation of lysine 27 on histone H3 in adult T-cell leukemia/lymphoma as a target for epigenetic therapy. Haematologica 2011;96:712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-κB pathway in adult T cell leukemia and other cancers. Cancer Cell 2012;21:121–35. [DOI] [PubMed] [Google Scholar]
- 14. Fujikawa D, Nakagawa S, Hori M, Kurokawa N, Soejima A, Nakano K, et al. Polycomb-dependent epigenetic landscape in adult T-cell leukemia. Blood 2016;127:1790–802. [DOI] [PubMed] [Google Scholar]
- 15. Ma G, Yasunaga JI, Shimura K, Takemoto K, Watanabe M, Amano M, et al. Human retroviral antisense mRNAs are retained in the nuclei of infected cells for viral persistence. Proc Nat Acad Sci U S A 2021;118:e2014783118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sugata K, Yasunaga J, Kinosada H, Mitobe Y, Furuta R, Mahgoub M, et al. HTLV-1 viral factor HBZ induces CCR4 to promote T-cell migration and proliferation. Cancer Res 2016;76:5068–79. [DOI] [PubMed] [Google Scholar]
- 17. Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2002;2:605–15. [DOI] [PubMed] [Google Scholar]
- 18. Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 1994;77:917–29. [DOI] [PubMed] [Google Scholar]
- 19. Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet 2000;26:464–9. [DOI] [PubMed] [Google Scholar]
- 20. Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 2008;22:2677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018;46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 2010;38:4958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakahata S, Ichikawa T, Maneesaay P, Saito Y, Nagai K, Tamura T, et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat Commun 2014;5:3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakagawa M, Shaffer AL 3rd, Ceribelli M, Zhang M, Wright G, Huang DW, et al. Targeting the HTLV-I-regulated BATF3/IRF4 transcriptional network in adult T cell leukemia/lymphoma. Cancer Cell 2018;34:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishio T, Kumar S, Shimono J, Daenthanasanmak A, Dubois S, Lin Y, et al. Genome-wide CRISPR screen identifies CDK6 as a therapeutic target in adult T-cell leukemia/lymphoma. Blood 2022;139:1541–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akkouche A, Moodad S, Hleihel R, Skayneh H, Chambeyron S, El Hajj H, et al. In vivo antagonistic role of the human T-cell leukemia virus type 1 regulatory proteins tax and HBZ. PLoS Pathog 2021;17:e1009219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamagishi M, Fujikawa D, Watanabe T, Uchimaru K. HTLV-1-mediated epigenetic pathway to adult T-cell leukemia-lymphoma. Front Microbiol 2018;9:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamagishi M, Hori M, Fujikawa D, Ohsugi T, Honma D, Adachi N, et al. Targeting excessive EZH1 and EZH2 activities for abnormal histone methylation and transcription network in malignant lymphomas. Cell Rep 2019;29:2321–37. [DOI] [PubMed] [Google Scholar]
- 30. Du W, Jiang P, Mancuso A, Stonestrom A, Brewer MD, Minn AJ, et al. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol 2013;15:991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le L, Li L, Li W, Chen T, Bin Z, Zhao L, et al. TAp73-induced phosphofructokinase-1 transcription promotes the Warburg effect and enhances cell proliferation. Nat Commun 2018;9:4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahgoub M, Yasunaga JI, Iwami S, Nakaoka S, Koizumi Y, Shimura K, et al. Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc Nat Acad Sci U S A 2018;115:E1269–e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamin D, Colombi M, Hindupur SK, Betz C, Lane HA, El-Shemerly MY, et al. Syrosingopine sensitizes cancer cells to killing by metformin. Sci Adv 2016;2:e1601756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benjamin D, Robay D, Hindupur SK, Pohlmann J, Colombi M, El-Shemerly MY, et al. Dual inhibition of the lactate transporters MCT1 and MCT4 is synthetic lethal with metformin due to NAD+ depletion in cancer cells. Cell Rep 2018;25:3047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11–20. [DOI] [PubMed] [Google Scholar]
- 36. Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev 2009;23:537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 2014;15:266–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rabinowitz JD, Enerbäck S. Lactate: the ugly duckling of energy metabolism. Nat Metab 2020;2:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gazon H, Chauhan PS, Porquet F, Hoffmann GB, Accolla R, Willems L. Epigenetic silencing of HTLV-1 expression by the HBZ RNA through interference with the basal transcription machinery. Blood Advances 2020;4:5574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sampath K, Ephrussi A. CncRNAs: RNAs with both coding and non-coding roles in development. Development 2016;143:1234–41. [DOI] [PubMed] [Google Scholar]
- 41. Hubé F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res 2010;39:513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol 2008;10:1098–105. [DOI] [PubMed] [Google Scholar]
- 43. Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep 2009;10:459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ong JZL, Yokomori R, Wong RWJ, Tan TK, Ueda R, Ishida T, et al. Requirement for TP73 and genetic alterations originating from its intragenic super-enhancer in adult T-cell leukemia. Leukemia 2022;36:2293–305. [DOI] [PubMed] [Google Scholar]
- 45. Kogure Y, Kameda T, Koya J, Yoshimitsu M, Nosaka K, Yasunaga JI, et al. Whole-genome landscape of adult T-cell leukemia/lymphoma. Blood 2022;139:967–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Engelmann D, Meier C, Alla V, Pützer BM. A balancing act: orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 2015;34:4287–99. [DOI] [PubMed] [Google Scholar]
- 47. Tanaka-Nakanishi A, Yasunaga J, Takai K, Matsuoka M. HTLV-1 bZIP factor suppresses apoptosis by attenuating the function of FoxO3a and altering its localization. Cancer Res 2014;74:188–200. [DOI] [PubMed] [Google Scholar]
- 48. Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 2019;47:W171–W4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishibashi R, Abe K, Ido N, Kitano S, Miyachi H, Toyoshima F. Genome editing with the donor plasmid equipped with synthetic crRNA-target sequence. Sci Rep 2020;10:14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Higuchi Y, Yasunaga JI, Mitagami Y, Tsukamoto H, Nakashima K, Ohshima K, et al. HTLV-1 induces T cell malignancy and inflammation by viral antisense factor-mediated modulation of the cytokine signaling. Proc Nat Acad Sci U S A 2020;117:13740–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bauer L, Müller LJ, Volkers SM, Heinrich F, Mashreghi MF, Ruppert C, et al. Follicular Helper-like T cells in the lung highlight a novel role of B cells in sarcoidosis. Am J Respir Crit Care Med 2021;204:1403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakagawa M, Schmitz R, Xiao W, Goldman CK, Xu W, Yang Y, et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med 2014;211:2497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoshida N, Shigemori K, Donaldson N, Trevisani C, Cordero NA, Stevenson KE, et al. Genomic landscape of young ATLL patients identifies frequent targetable CD28 fusions. Blood 2020;135:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka A, Ishitsuka Y, Ohta H, Fujimoto A, Yasunaga JI, Matsuoka M. Systematic clustering algorithm for chromatin accessibility data and its application to hematopoietic cells. PLoS Comput Biol 2020;16:e1008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022;38:2943–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vera Alvarez R, Pongor LS, Mariño-Ramírez L, Landsman D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 2019;35:1960–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 2019;9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Limbach M, Saare M, Tserel L, Kisand K, Eglit T, Sauer S, et al. Epigenetic profiling in CD4+ and CD8+ T cells from Graves’ disease patients reveals changes in genes associated with T cell receptor signaling. J Autoimmun 2016;67:46–56. [DOI] [PubMed] [Google Scholar]
- 70. Wong RWJ, Ngoc PCT, Leong WZ, Yam AWY, Zhang T, Asamitsu K, et al. Enhancer profiling identifies critical cancer genes and characterizes cell identity in adult T-cell leukemia. Blood 2017;130:2326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 2010;28:1045–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 2015;350:334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zou Z, Ohta T, Miura F, Oki S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res 2022;50:W175–W82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iwata A, Durai V, Tussiwand R, Briseño CG, Wu X, Grajales-Reyes GE, et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat Immunol 2017;18:563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next-generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556–W60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kawano Y, Sasano T, Arima Y, Kushima S, Tsujita K, Matsuoka M, et al. A novel PDK1 inhibitor, JX06, inhibits glycolysis and induces apoptosis in multiple myeloma cells. Biochem Biophys Res Commun 2022;587:153–9. [DOI] [PubMed] [Google Scholar]
- 78. Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Human cluster 2 genes whose H3K27me3 in TSS's was decreased.
Mouse cluster 2 genes whose H3K27me3 in TSS's was decreased.
Oligonucleotides used in this study.
Experimental reagents and resources with identifiers used in this study.
Transcriptional regulation by HBZ protein and RNA and the status of TP73 expression.
HBZ protein alters epigenetic regulation of TAp73 by EZH2.
Regulation of TP73 by BATF3 and IRF4.
TAp73 significance in HBZ pathogenesis and its effect on cellular metabolism.
Transcriptional regulation of SLC16A1 and SLC16A3 by TAp73, and mRNA expression levels of MCT-coding genes among various cancers.
Efficacy of the MCT1/4 inhibitor syrosingopine on ATL cells.
Data Availability Statement
Sequencing data in this study have been deposited with links to BioProject accession number PRJDB15617 in the DDBJ BioProject database.



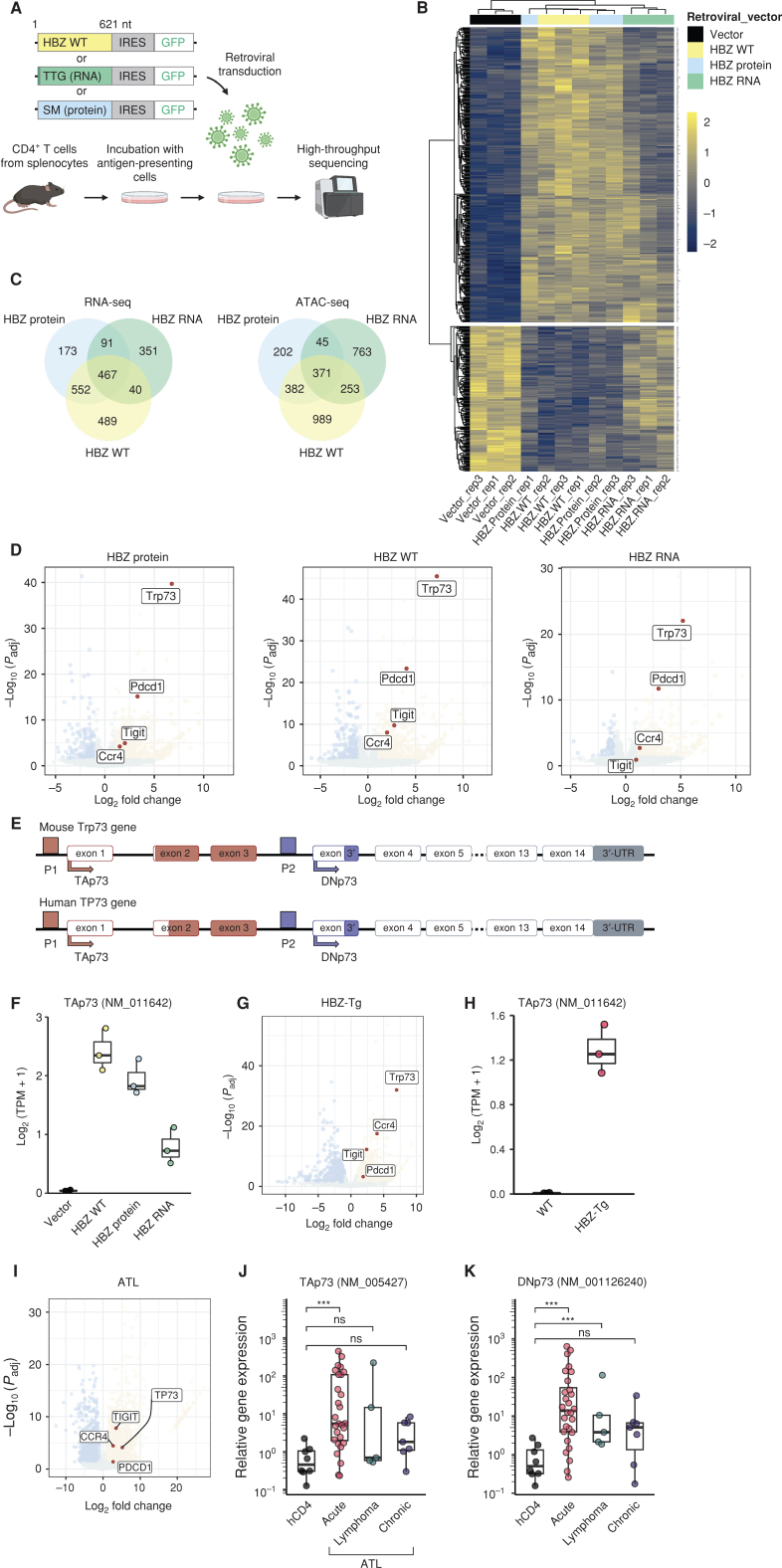
![Figure 2. HBZ protein alters EZH2 genome-wide distribution and decreases its binding to the TAp73 promoter. A, GSEA plots for mouse CD4+ T cells transduced with WT or mutant HBZ compared with the vector. The normalized enrichment score (NES) and q-value are listed. B, Immunoprecipitation (IP) with anti-Flag antibody (Flag-HBZ) showing interaction between HBZ protein and EZH2 in HEK293T cells. IP was analyzed by SDS-PAGE and immunoblotting (IB). C–E, ChIP-qPCR for EZH2 and H3K27me3 in the TAp73 promoter region. The %Input is shown for WT or HBZ-Tg mouse CD4+ T cells (C), human CD4+ T cells from healthy donors (hCD4) or ATL cell lines (D), and Jurkat cells with stable transduction of WT or mutant HBZ (E; n = 3). F, Histone methyltransferase (HMT) activity on H3K27 among transduced Jurkat cells (n = 3). G and H, Heat maps for genomic regions with enriched ChIP-seq scores for H3K27me3. The score for each region was scaled and clustered based on healthy human donor CD4+ T cells (hCD4; G) or WT mouse CD4+ T cells (H) with transcription start site (TSS) and transcription end site (TES) labeling. In addition to Treg, ATL cells (TL-Om1 and ED; G) and cells from HBZ-Tg mice (H) were analyzed. I, Representative genes found in cluster 2 that were shared between the results from human and mouse cells in G and H, respectively. J, Results of KEGG pathway analysis using the genes in cluster 2 for humans (G). Shared pathways between human and mouse cells are highlighted in red. Statistical values and gene counts calculated by the clusterProfiler are shown. K and L, Scatter plots of mouse cluster 2 genes resulting from combinational analysis of RNA-seq (HBZ-Tg mouse CD4+ T cells relative to WT) and ChIP-seq [relative enrichments in TSS of WT as area under the curve (AUC)] for EZH2 (K) and H3K27me3 (L). Results are plotted as mean ± SD, using Student t test (C) or one-way ANOVA with the post hoc Dunnet test (D–F). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c3de/10473166/b2b2d7ad393d/374fig2.jpg)