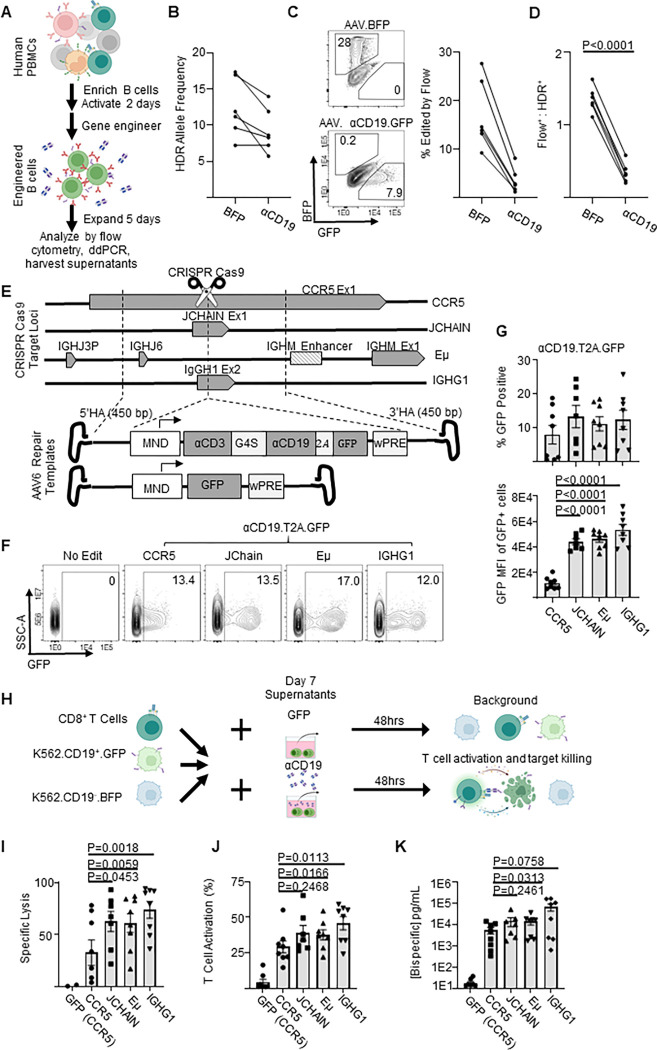
Figure 1: Genome engineered primary human B cells secrete functional αCD19-bispecific in a locus dependent manner.
A) Schematic showing the experimental flow of a primary B cell experiment. Briefly, after isolation from PBMCs, B cells were edited to express either BFP or αCD19.T2A.GFP transgenes at CCR5 genetic loci via HDR-gene editing with AAV6 delivered DNA repair templates. Five days later genomic DNA, cells and supernatants were analyzed as indicated. B) Transgene integration at CCR5 locus shown here as HDR allele frequency was measured by ddPCR. C) Representative flow cytometry plots showing transgene expression of fluorescent proteins in engineered B cells shown and quantified as % edited of live cells. D) Ratio of engineering rate as determined by ddPCR vs flow cytometry. E) Schematic showing the editing strategies for delivery of GFP or αCD19.T2A.GFP to antibody-associated loci. F) Representative flow cytometry plots of αCD19.T2A.GFP edited B cells with G) the quantification of % edited and GFP mean fluorescent intensity of edited cells. H) K562 killing assay schema. Supernatants from edited B cells were incubated with target (CD19+) and control (CD19−) K562 cells with CD8+ T cells for 48 hours. Cells were harvested for flow cytometry to obtain I) specific lysis of CD19+ K562 and J) T cell activation (%CD69+CD137+ of CD3+ cells). K) The concentration of bispecific in the supernatants as interpolated from %T cell activated data. Data are from five donors in five independent experiments. Error bars represent SEM. P values calculated using D) a paired student’s t test and G, I-K) paired one-way ANOVAs with Dunnett’s posttest. Illustrations were created in part with biorender.com.