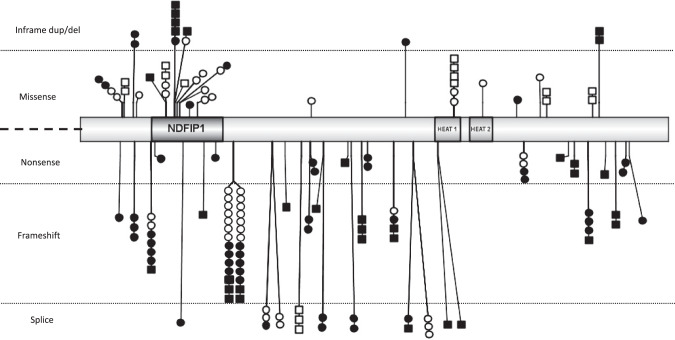
Fig. 3. Graphical representation of BRAT1 variants (IBS 1.0.3).
Missense and in-frame deletions are above; nonsense, frameshift and splice variants are below. Variants in black were identified in patients with RMFSL phenotype, those in white in patients with NEDCAS. Each square represents a patient with a homozygous variant, each circle a patient with a heterozygous one. Note that all the patients with a homozygous loss-of function variant are presenting a RMFSL phenotype and that the only one patient with a homozygous missense variant and a severe phenotype may have a second associated etiology. NDFIP1, NEDD4 family interacting protein 1; HEAT1/HEAT2, protein tandem repeat structural motif found in the four proteins: Huntingtin, Elongation factor 3 (EF3), protein phosphatase 2 A (PP2A) and TOR1.