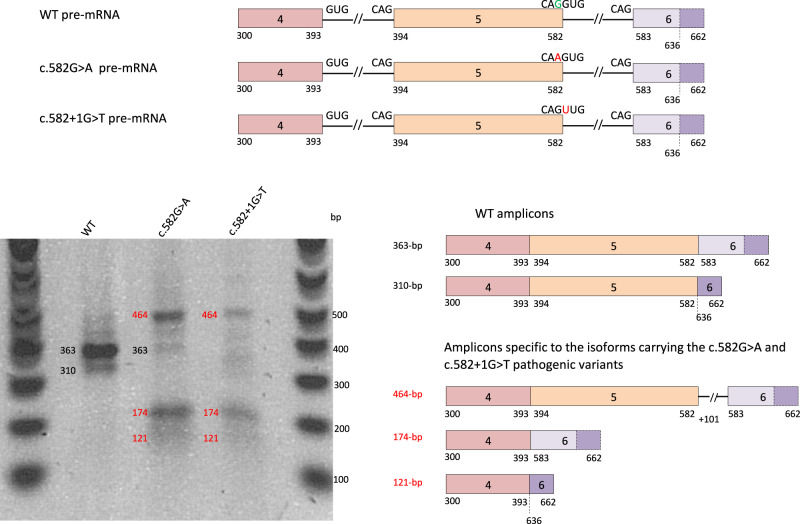
Fig. 2. In vitro assessment of SFTPB splicing defects associated with c.582G>A p.(Gln194=).
SFTPB exons 4–6 pre-mRNA corresponding to plasmids pSFTPB-WT, pSFTPB_mut and pSFTPB_ctr are represented. The reference nucleotide is in green and the pathogenic variant is in red. The dotted line highlights a cryptic acceptor splice site in exon 6 (nucleotide 636). After transfection of A549 cells with the above-mentioned plasmids, the migration on a 1.5% agarose-BET gel of the RT-PCR products obtained from extracted RNAs is shown. The main amplicons characterized by Sanger sequencing and obtained from the normal and mutant constructs are represented on the right of the gel image. Two amplicons were observed for the pSFTPB_WT: a 363 bp (canonical splicing) and a 310 bp (using exon 6 cryptic splice-acceptor site corresponding to putative p.(Asp195Glyfs*39)). Both the c.582G>A (pSFTPB_mut) and the c.582+1G>T (pSFTPB_ctr) pathogenic variants resulted in three specific and aberrant amplicons, i.e., from top to bottom: one 464 bp transcript related to the retention of the first 101 bp of intron 5 and predicting a frameshift with a premature Stop codon in exon 6 (p.(Asp195Valfs*54)), and two 174 and 121 bp amplicons lacking exon 5 and corresponding to the use of intron 5 canonical splice-acceptor site (p.(Asp132_Gln194del) in-phase deletion), or exon 6 cryptic splice-acceptor site (p.(Asp132Glyfs*39)) respectively. The data are representative from 3 independent experiments. WT wild type.