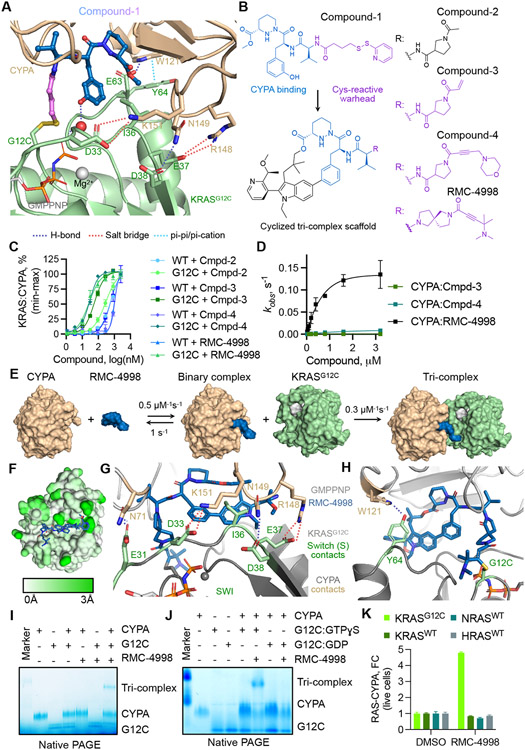
Fig. 1. Development of a tri-complex inhibitor targeting the active state of KRASG12C.
(A) Tertiary structure of the primitive CYPA:Compound-1:KRASG12C-GMPPNP complex at 1.40 Å resolution. (B) The chemical structure of Compound-1 and the macrocyclized scaffold for subsequent compounds, with the sanglifehrin-derived CYPA-binding moiety shown in blue. R denotes divergent chemical moieties comprising non-covalent (black) and covalent (pink) derivatives. (C) The interaction between the indicated KRAS proteins (50 nM) and CYPA (20 μM) in the presence of increasing compound concentrations was determined by TR-FRET (mean ± SEM, n=3, N=2). (D) The rate (kobs) of covalent modification of KRASG12C in the presence of CYPA (25 μM) and the indicated compounds (mean ± SEM, N=3). (E) Schematic of tri-complex formation by RMC-4998 along with rate constants for each reaction. (F) Mapped pairwise atomic distances between structures of apo-CYPA (PDB: 3K0N) and RMC-4998-bound CYPA showing the structural rearrangements (white to green gradient) that gave rise to the high-affinity neomorphic binding interface for KRASG12C. Interactions between RMC-4998, CYPA, and the Switch I (G) or Switch II (H) regions of KRASG12C in the crystal structure of the mature tri-complex (1.53 Å). (I) The interaction between KRASG12C (2 μM), RMC-4998 (10 μM), and CYPA (2 μM) was determined by native PAGE. (J) As in J, but KRASG12C was loaded with non-hydrolysable GTPɣS (active state) or GDP (inactive state). A representative of at least two independent experiments is shown in (I) and (J). (K) HEK293 cells co-expressing small bit luciferase-tagged RAS variants and large-bit luciferase tagged CYPA were treated with RMC-4998 (100 nM) for 2h followed by determination of luciferase activity (mean ± SEM, N=4).