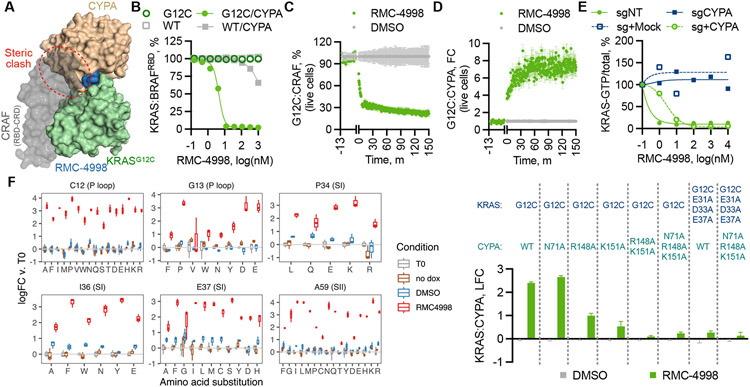
Fig. 2. Structural constraints for tri-complex formation and active state selective KRAS inhibition.
(A) Superimposition of the structures of the CYPA:RMC-4998: KRASG12C tri-complex and the KRAS:CRAFRBD/CRD complex (PDB: 6XI7) (31). (B) The effect of RMC-4998 on the interaction between the indicated KRAS proteins (12.5 nM) and BRAFRBD (50 nM) in either the presence or the absence of CYPA was determined by TR-FRET (mean ± SD, N=4). HEK293 cells co-expressing small bit luciferase-tagged KRASG12C and large bit luciferase-tagged CYPA (C) or full-length CRAF (D) were treated with RMC-4998 (100 nM) followed by determination of reconstituted luciferase activity in live cells (mean ± SEM, N=3). (E) The indicated parental, CYPA-null or CYPA rescued H358 cells were treated as shown and their extracts were analyzed by RBD-pulldown and immunoblotting to determine the effect on KRAS activation. A representative of two independent experiments is shown. (F) KRAS mutant cells were infected with a dox-inducible saturation mutagenesis library based on a KRASG12C backbone and treated with either DMSO or RMC-4885 (100 nM) for two weeks. Shown is the log(fold-change) (logFC) in abundance relative to T0 (mean ± 95%CI, N=3) for variants meeting the threshold for statistical significance (see Methods). (G) The effect of the indicated CYPA variants on tricomplex formation in live cells was determined as in C (mean ± SEM, N=3).