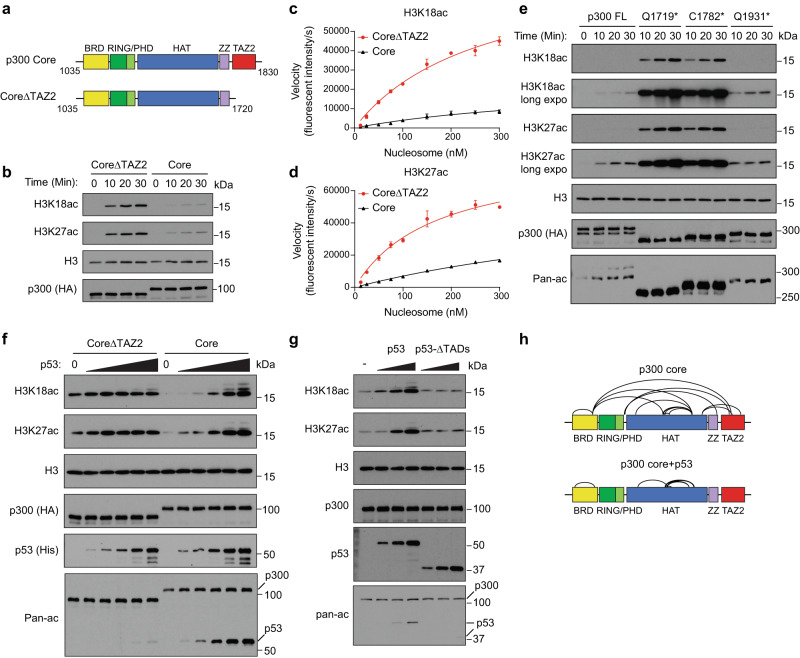
Fig. 3. The TAZ2 domain negatively regulates p300 HAT activity.
a Schematic of p300 Core and CoreΔTAZ2. Domains are color coded as in Fig. 1a. b Western blots of in vitro HAT assays using p300 Core and CoreΔTAZ2 proteins purified from 293 T cells. c, d Kinetic measurement of the catalytic activity of p300 Core and CoreΔTAZ2 on nucleosome. The steady-state velocity was determined based on fluorescent intensity of H3K18ac c and H3K27ac d per second. The Km of nucleosome substrate (derived from non-linear Michaelis-Menten regression) by Core and CoreΔTAZ2 are 0.82 ± 0.23 μM and 0.17 ± 0.03 μM for H3K18ac, and 0.50 ± 0.14 μM and 0.27 ± 0.04 μM for H3K27ac, respectively. Error bars represent standard error of the mean (s.e.m.) of 3 biological replicates. Representative quantitative Western blots are shown in Supplementary Fig. 5e. e Western blots of in vitro HAT assays using the purified full-length p300 protein and truncation mutants identified in human cancers. f Western blots of in vitro HAT assays of p300 Core and CoreΔTAZ2 proteins in the presence of increasing amounts of p53 protein. g Western blots of in vitro HAT assays of p300 Core in the presence of full-length p53 or p53ΔTADs. Western blots are representative data of 3 b, e, g or 2 f biological experiments. h Crosslinks (black arcs) identified by CLMS of p300 Core with and without p53 incubation (details in Supplementary Data 6). Source data are provided as a Source Data file.