Fig. 7. Signaling pathways involved in the regulatory effect of FAM3A on VSMC differentiation reprogramming.
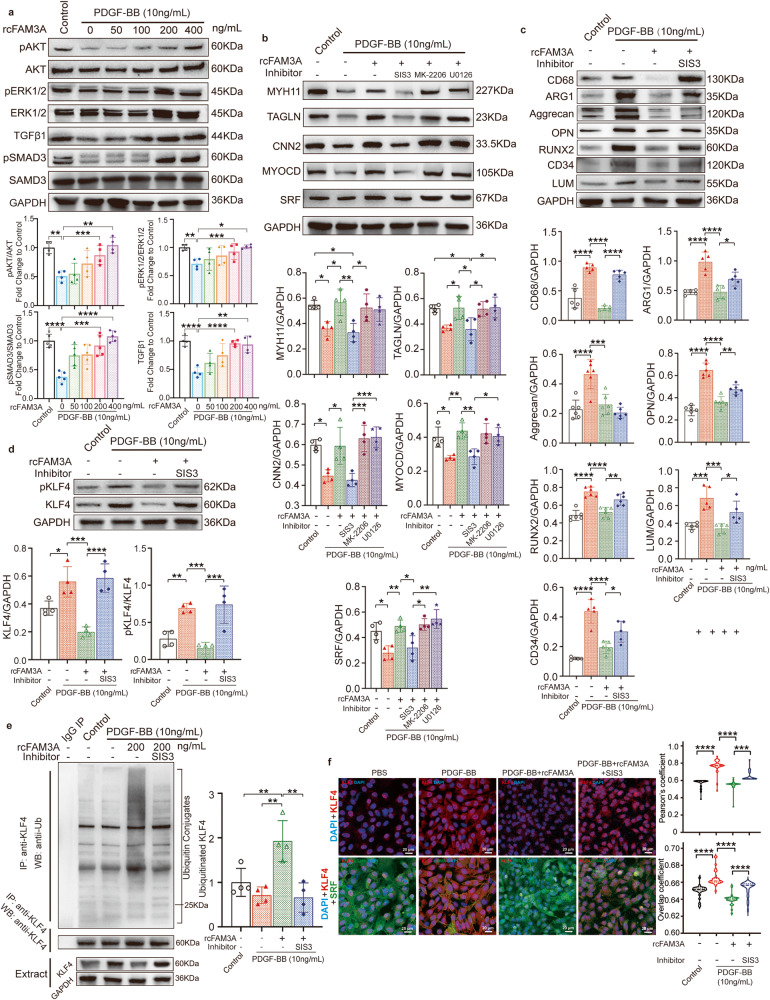
a VSMCs were stimulated with PDGF, and further treated with or without recombinant FAM3A at the indicated concentration gradient. The AKT, ERK1/2, and TGFβ/SMAD3 pathways in whole cell lysates were detected by western blots. Representative images and quantification are shown (n = 4 biologically independent experiments). b–d VSMCs were stimulated with PDGF and recombinant FAM3A (200 ng/ml) with or without the inhibitors of the TGFβ/SMAD3 (SIS3), AKT (MK-2206), or ERK1/2 (U0126) pathways as indicated in the charts. Representative western blot images and quantification of VSMC contractile markers (b, n = 4 biologically independent experiments), VSMC transdifferentiation markers (c, n = 5 biologically independent experiments), or phosphorylated KLF4 (Ser254) (d, n = 4 biologically independent experiments) in whole cell lysates are shown. e Cells were treated as in (c). Whole cell lysates were prepared and KLF4 protein was precipitated with anti-KLF4. The presence of ubiquitin-conjugated KLF4 in the immunocomplex was detected by western blot with anti-Ub (upper). The amount of precipitated KLF4 in the immunocomplex was determined by anti-KLF4 (middle). The KLF4 and GAPDH (lower) were detected in the same whole cell lysates (Extract). Representative western blot images and quantification of KLF4 ubiquitination levels normalized to GAPDH are shown (n = 4 biologically independent experiments). Quantitative comparisons between samples were run on the same gel (a–e). f VSMCs were treated as in (c). Confocal imaging showed the cellular localization of nuclei (blue), KLF4 (red), and SRF (green). The colocalization of nuclei and KLF4 was analyzed by ImageJ software with Pearson’s correlation coefficient (PCC, upper) and Manders’ colocalization coefficients (MCC, lower) quantifications (n = 4 biologically independent experiments with 24 scans for each group). Scale bar: 20 μm. Data are presented as mean ± SEM (a–e) and median ± IQR (f). Statistical significance was calculated with one-way ANOVA followed by Tukey post hoc test (a–e) and Kruskal–Wallis test followed by Nemenyi post hoc test (f) and P values are indicated (*P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Source data are provided as a Source Data file.