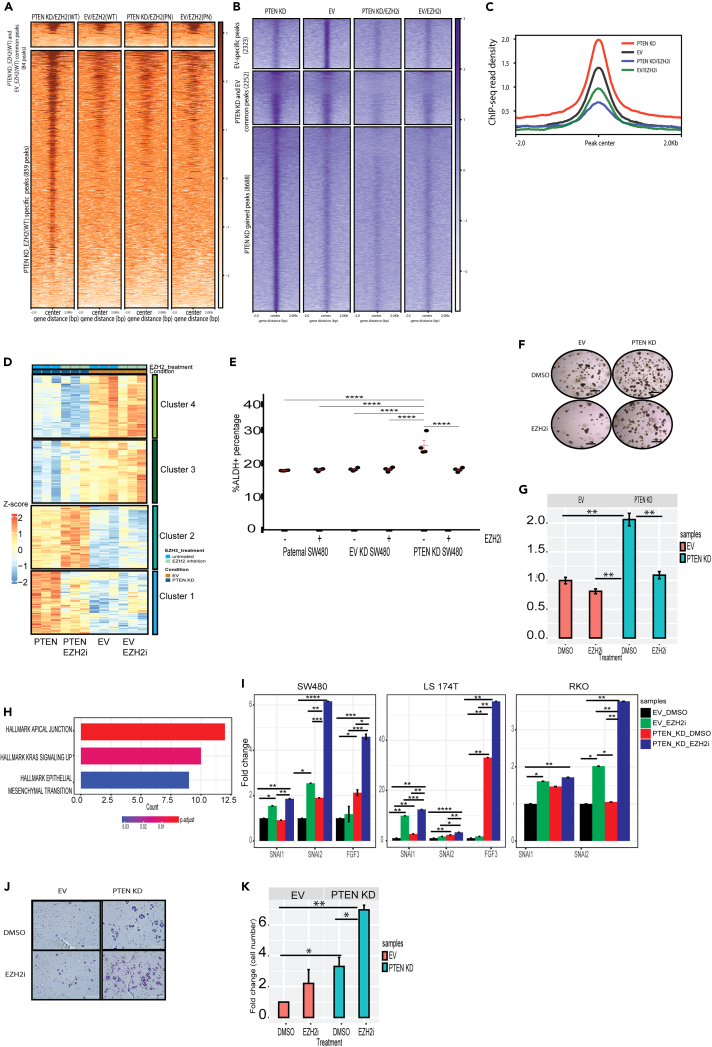
Figure 7.
EZH2 inhibition blocks the PTEN knockdown-induced increase in β-catenin enrichment over the genome
(A) Metagenomic heatmap for HA CUT&RUN prepared from HA-EZH2 wildtype (WT) and HA-EZH2 S21A expressing PTEN knockdown (KD) and empty vector (EV) SW480 cells.
(B) Metagenomic heatmap for FLAG ChIP-seq prepared from FLAG-β-catenin WT and FLAG-β-catenin K49R expressing PTEN KD and EV expressing SW480 cells.
(C) Average ChIP-seq read intensity for all peaks shown in B.
(D) Heatmap for differentially expressed genes (DEGs) in PTEN KD versus EV clustered manually based on the effect of EZH2 inhibitor (GSK503, 1 μM, 72 h).
(E) ALDEFLUOR assay showing the percentage of ALDH+ cells in cells treated as in D. Red lines indicate mean +/− SEM.
(F) Brightfield images of spheroids following treatment as in D. Scale bar = 2000 microns.
(G) Quantification of spheroids normalized to spheroids counts for untreated EV cells. Results are represented as mean of 3 biological replicates +/− SEM.
(H) Barplot for the hallmark analysis of DEGs in cluster 2.
(I) Gene expression of the indicated genes by RT-qPCR in SW480, LS174T and RKO cells treated as in D. Expression of all the genes was normalized to the house keeping gene RHOA and then to untreated EV cells. Results are represented as mean of 3 biological replicates +/− SEM.
(J) EV and PTEN KD cells were treated with DMSO or 2 μM EZH2 inhibitor (EZH2i, GSK-503) for 72 h followed by plating cells in the upper chamber of a transwell insert. Brightfield images of crystal violet–stained migrated cells were taken after 48 h. Scale bar = 200 microns.
(K) Quantification of migration normalized to migration counts for untreated EV cells. Results are represented as mean of 3 biological replicates +SEM. Significance was determined by one-way ANOVA with the Tukey multiple comparisons test. All significant comparisons are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figures S6 and S7.