Abstract
Purpose
Association between variants rs1047972 and rs8173 of the AURKA gene in healthy women and breast cancer (BC) in a Mexican population.
Methods
Genomic DNA samples from 409 healthy women and 572 patients with BC were analyzed for variants rs1047972 and rs8173 of the AURKA gene by polymerase chain reaction-restriction fragment length polymorphism.
Results
TT genotype (odds ratio [OR], 2.5; 95% confidence interval [CI], 1.22–5.11; p = 0.0015) and the T allele (OR, 1.16; 95% CI, 1.23–2.12; p = 0.0007) of the rs1047972 variant were associated as risk susceptibility for BC relative to the control group. Contrarily, the GG genotype (OR, 0.64; 95% CI, 0.43–0.94; p = 0.029) was associated as a protective factor of susceptibility of BC of the variant rs8173 of the AURKA gene. Differences were observed in the patients with BC who were carriers of the CT genotype of the rs1047972 variant with overweight, obesity, estrogen receptor-positive plus obesity, Ki-67 (≥ 20%) plus history familial positive of cancer; and for variant rs8173 the BC patients who were CG carriers and presented chemotherapy gastric toxicity, hormonal receptor positive plus chemotherapy gastric toxicity, and menopause status plus chemotherapy gastric toxicity (p < 0.05). Two common haplotypes were identified in the study groups: CG and TC genotypes, were associated as a protective and risk factor, respectively (p < 0.05).
Conclusion
Variants rs1047972 and rs8173 of the AURKA gene and the TC haplotype were associated as risk susceptibility factors for BC in this population.
Keywords: Aurora Kinase A, Breast Neoplasms, DNA Restriction Enzymes, Genotype, Risk Factors
INTRODUCTION
Breast cancer (BC) is the most frequently diagnosed malignancy and the primary cause of death among women in Mexico and the world [1,2]. According to data provided by the Global Cancer Observatory (GLOBOCAN), it is estimated that up to eight million cases were diagnosed and nearly two million new cases were reported in 2020, along with 684,996 deaths, representing the type of cancer with the highest prevalence, incidence, and mortality among women worldwide [1,2].
In Mexico, this disease is reported as the neoplasia with the highest prevalence, incidence, and mortality, with just under thirty thousand new cases diagnosed in 2020 and nearly eight thousand deaths reported during that year (GLOBOCAN) [1]. Previous studies have shown that BC represents a public health problem, which some developed countries have managed to face and drastically reduce mortality rates by implementing new advances in the diagnosis and treatment of this disease; however, this is not the case in underdeveloped countries, where an increase in mortality rates has been observed [3,4].
Although BC is a multifactorial disease, the genetic component plays an important role in the appearance and behavior of this pathology; therefore, the study of variants and especially single nucleotide variants (SNV) in genes involved in the carcinogenesis process is of particular interest. Studying SNVs makes it possible to understand the genomic changes that may increase the susceptibility to developing this pathology, or influence the therapeutic response, and contribute to the clinicopathological characteristics making this type of cancer a very heterogeneous disease [5,6,7].
One of the genes of interest is the Aurora kinase (AURK) A (AURKA) gene located in 20q13.2, which is part of the AURK family, whose members, the AURKA, AURK B (AURKB), and AURK C (AURKC) genes, play an important role in the formation of the mitotic spindle, and consequently, an important role in cell division by guaranteeing correct chromosome segregation, therefore, defects in any of the genes of this family can promote genetic instability and polyploidies, characteristics already described in numerous neoplasms [8,9].
The AURKA gene plays an important role in numerous mitotic activities, such as centrosome maturation, progression to mitosis, and mitotic spindle assembly, through coupling with other proteins, thus ensuring the phosphorylation of key proteins in the regulation of the cell cycle, including P53 and breast cancer gene (BRCA) 1 [10,11]. Thus, the deregulation of this gene being directly related to carcinogenic processes is unsurprising. Furthermore, AURKA is overexpressed in numerous tumor tissues and the overexpression of this gene in breast cancer is related to aggressive highly proliferative tumors, with basal phenotypes and Ki-67 positivity [8]. Similarly, the overexpression of this gene plays an important role as a prognostic marker in adrenocortical carcinoma, clear and papillary renal cell carcinoma, hepatocellular carcinoma, lung adenocarcinoma, mesothelioma, and uveal melanoma. Thus, the AURKA gene is considered a therapeutic target of great importance in precision medicine for identifying variants that may influence the development, prognosis, therapeutic response, and progression of the disease [12,13,14,15,16].
Variants of this gene have been associated with the appearance, development, and poor prognosis of different types of cancer. For example, the rs2273535 variant is associated with the appearance of multiple types of cancer, especially BC. Additionally, the variants rs2064863 and rs6024836 have been reported to be associated with different clinicopathological characteristics in patients with urothelial cell carcinoma. The rs1047972 variant has also been correlated with high rates of hepatocellular carcinoma [17,18,19].
The rs1047972 variant is a non-synonymous SNV that mediates the substitution of a valine residue for an isoleucine residue at position 57 (V57I) [20] and has been extensively studied. However, there have been contradictory results in some studies showing protective factors, especially in BC [21,22], others as risk factors [23,24,25], and others have not been shown to be associated [26]. However, the three prime untranslated region (3′UTR) region of the AURKA gene plays an important role in the miRNA expression in the tumor microenvironment [27]. Additionally, miR-124 acts as a tumor suppressor by significantly decreasing the expression of AURKA in a glioma cell line [28]. Thus, the presence of SNVs in the 3′UTR region of numerous genes modify the binding sites to regulatory miRNAs, which is associated with different human pathologies and the therapeutic response of some of them, including cancer [12].
Although SNVs rs1047972 and rs8173 have been extensively studied in different populations, no association studies have been reported for BC or any other type of cancer in the Mexican population. Therefore, in this study, we analyzed the association of the variants rs1047972 (T>C) and rs8173 (G>C), located in exon 3 and the 3′UTR region of the AURKA gene, respectively.
METHODS
The present study was approved by the local ethics committee (1305) of the “Centro de Investigación Biomédica de Occidente, Instituto Mexicano del Seguro Social”. DNA samples from 572 women with clinically and histologically confirmed BC, and 409 healthy women were used in this study. Sample collection was performed after written informed consent was provided by each subject in addition to obtaining clinical data. All the study procedures were performed in accordance with the Declaration of Helsinki.
The rs1047972 variant was tested in 304 patients with BC and 272 controls. The polymerase chain reaction (PCR) amplification was carried out using the forward 5′-CTCTTCCATTCTAGGCTAC-3′ and reverse 5′-CAGTGGCCTGGAGACAG-3′ primers designed according to the gene sequence (http://www.ncbi.nlm.nih.gov/SNP/). A fragment of 247 bp was amplified with the following program: 95°C for 5 minutes; 35 cycles of 95°C for 30 seconds, 62°C for 45 seconds, and 72°C for 30 seconds. The genotype identification was performed by restriction fragment length polymorphism (RFLP) method, using the restriction enzyme BstUI (New England Biolabs, Ipswich, USA) that recognizes the G allele. In a polyacrylamide gel 8% (19:1), the fragment was separated by electrophoresis after staining with silver nitrate. The CC genotype was identified by a band of 247 bp, the GT by bands of 247, 139, and 108 bp, and the GG genotype by bands of 139 and 108 bp.
The rs8173 variant was analyzed in 572 patients with BC and 409 controls. A fragment of 184 bp was amplified by PCR with the forward 5′-GTGCCGGACAGACACACAG-3′ and reverse 5′-GGAGCTGTGCAATAACC-3′ primers designed according to the sequence of the gene (http://www.ncbi.nlm.nih.gov/SNP/). Using the 35 cycles of alignment temperature at 59°C for 5 seconds, 57°C for 5 seconds, 55°C for 20 seconds, and 72°C for 30 seconds. Genotypes were identified prior to digestion with the restriction enzyme HaeIII (New England Biolabs, Ipswich, USA) and the fragments were separated on 8% polyacrylamide gels (19:1) after staining with silver nitrate. The following fragments were used to identify the genotypes: CC, 184 bp; CG, 184, 136, and 48 bp; and GG 136 and 48 bp.
Statistical analysis
The percentage of genotypes was analyzed by direct count, and Hardy-Weinberg equilibrium (HWE) was used for the control group using the χ2 goodness-of-fit test to compare the observed genotype frequencies with the expected frequencies. For genotype association analysis, odds ratios were calculated, and binary logistic regression was performed using SPSS Statistics Base 24 software (IBM, Chicago, USA). The SHEsis Online Version program was used to analyze pairwise linkage disequilibrium (D′) and haplotype frequency [29]. The allelic frequencies from other populations were taken from the Ensembl database using the reference genome “CRCh38.p13.”
RESULTS
The average age observed in the study groups was 52.02 ± 11.72 and 51.78 ± 11.65 years for the BC and control groups, respectively, which did not show statistically significant differences when compared to each other (p > 0.05). Moreover, the comparative analysis of tobacco and alcohol consumption showed no significant differences between the study groups (p > 0.05).
The frequencies of clinical variables in the BC group are shown in Table 1. Among the most frequent variables were, the presence of menarche between 11 and 13 years old (69%), menopause (62%), family history of cancer (44%), obesity (47%), less than five gestations (68%), breastfeeding more than 6 months (50%), location of cancer in the unilateral breast (96%), ductal type (92%); stage II (32%), and III (30%), positive metastatic nodes (72%); luminal A 39%, and luminal B (25%), no responders (60%), and gastric toxicity (62%) to chemotherapy.
Table 1. Clinical data for the breast cancer group.
| Variables | BC patients (n = 572) | |
|---|---|---|
| Menarche status | ||
| 7 at 10 years old | 52 (9.1) | |
| 11 at 13 years old | 396 (69.2) | |
| 14 at 18 years old | 124 (21.7) | |
| Menopause status | ||
| Premenopausal | 216 (37.8) | |
| Postmenopausal | 356 (62.2) | |
| Family history of cancer | ||
| Breast* | 130 (22.7) | |
| Other type* | 118 (20.6) | |
| Body mass index (kg/m2) | ||
| 18–24.9 (normal weight) | 154 (26.9) | |
| 25–29.9 (overweight) | 149 (26.0) | |
| ≥ 30 (obesity) | 269 (47.0) | |
| Hormonal consumption* | 218 (38.1) | |
| Pregnancies status | ||
| ≤ 4 | 390 (68.2) | |
| ≥ 5 | 121 (21.2) | |
| nulliparity | 61 (10.7) | |
| Miscarriage* | 133 (23.3) | |
| Breastfeeding (months) | ||
| ≤ 6 | 213 (37.2) | |
| > 6 | 288 (50.3) | |
| No | 71 (12.4) | |
| Localization (breast left or right) | ||
| Unilaterality | 547 (95.6) | |
| Bilateral | 25 (4.4) | |
| Histology (adenocarcinoma) | ||
| Ductal | 526 (92.0) | |
| Lobular | 35 (6.1) | |
| Mixed | 11 (1.9) | |
| Tumor stage | ||
| I | 63 (11.0) | |
| II | 183 (32.0) | |
| III | 172 (30.1) | |
| IV | 154 (26.9) | |
| Lymph node status | ||
| Positive* | 413 (72.2) | |
| Molecular type | ||
| Luminal A | 224 (39.2) | |
| Luminal B | 145 (25.3) | |
| Her-2 | 93 (16.3) | |
| Triple negative | 110 (19.2) | |
| Ki-67 (≥ 20%) | 284 (49.7) | |
| Ki-67 (< 20%) | 288 (50.3) | |
| Chemotherapy status | ||
| Response | 230 (40.2) | |
| No response | 342 (59.8) | |
| Personal medical history | ||
| Benign breast disease- uterine fibroids* | 178 (31.1) | |
| DM2-hypertension* | 132 (23.1) | |
| Chemotherapy toxicity | ||
| Gastric | 355 (62.1) | |
| Hematologic | 217 (37.9) | |
Values are presented as number of patients (%).
BC = breast cancer; DM2 = type 2 diabetes mellitus.
*On base 572.
In the rs1047972 variant the TT genotype frequency, (odds ratio [OR], 2.5; 95% confidence interval [CI], 1.22–5.11; p = 0.015), T allele (OR, 1.6; 95% CI, 1.23–2.12; p = 0.0007), dominant model (OR, 1.6; 95% CI, 1.18–2.29; p = 0.003), and recessive model (OR, 2.5; 95% CI, 1.22–5.11; p = 0.015) were observed as risk factors for BC. However, in the rs8173 variant, the GG genotype frequency, and recessive model, (OR, 0.64; 95% CI, 0.43–0.94; p = 0.029), were observed as a protective factor for BC (Table 2).
Table 2. Genotype and allelic distribution of the rs1047972, and rs8173 variants of AURKA gene in the studied groups.
| Variants | BC | Controls* | OR (95% CI) | p-value | |||
|---|---|---|---|---|---|---|---|
| rs1047972 | |||||||
| Genotypes | n = 304 | n = 272 | |||||
| CC | 159 (52.3) | 175 (64.3) | 1.0 | ||||
| CT | 116 (38.2) | 86 (31.6) | 1.3 (0.94–1.88) | 0.1 | |||
| TT | 29 (9.5) | 11 (4.0) | 2.5 (1.22–5.11) | 0.015 | |||
| Dominant | |||||||
| CC | 159 (52.3) | 175 (64.3) | |||||
| CT+TT | 145 (47.7) | 97 (35.7) | 1.6 (1.18–2.29) | 0.003 | |||
| Recessive | |||||||
| TT | 29 (9.5) | 11 (4.0) | 2.5 (1.22–5.11) | 0.015 | |||
| CC+CT | 275 (90.5) | 261 (96.0) | |||||
| Alleles | 2n = 608 | 2n = 544 | |||||
| C | 434 (71.4) | 436 (80.1) | 0.61 (0.46–0.81) | 0.0007 | |||
| T | 174 (28.6) | 108 (19.9) | 1.6 (1.23–2.12) | 0.0007 | |||
| rs8173 | |||||||
| Genotypes | n = 572 | n = 409 | |||||
| CC | 202 (35.3) | 145 (35.5) | 1.0 | ||||
| CG | 311 (54.4) | 202 (49.4) | 1.22 (0.94–1.57) | 0.140 | |||
| GG | 59 (10.3) | 62 (15.2) | 0.64 (0.43–0.94) | 0.029 | |||
| Dominant | |||||||
| CC | 202 (35.3) | 145 (35.5) | |||||
| CG+GG | 370 (64.7) | 264 (64.5) | 1.0 (0.77–1.31) | 0.964 | |||
| Recessive | |||||||
| GG | 59 (10.3) | 62 (15.2) | 0.64 (0.43–0.94) | 0.029 | |||
| CC+CG | 513 (89.7) | 347 (84.8) | |||||
| Alleles | 2n = 1,144 | 2n = 818 | |||||
| C | 715 (62.5) | 492 (60.1) | 1.10 (0.91–1.32) | 0.312 | |||
| G | 429 (37.5) | 326 (39.9) | 0.90 (0.75–1.08) | 0.312 | |||
p-value less than 0.05 considered statistically significant.
BC = breast cancer; OR = odds ratio; CI = confidence interval.
*Hardy-Weinberg equilibrium for the control group: variants rs1047972 (χ2 test = 0.0113, p = 0.915), and rs8173 (χ2 test = 0.373; p = 0.541).
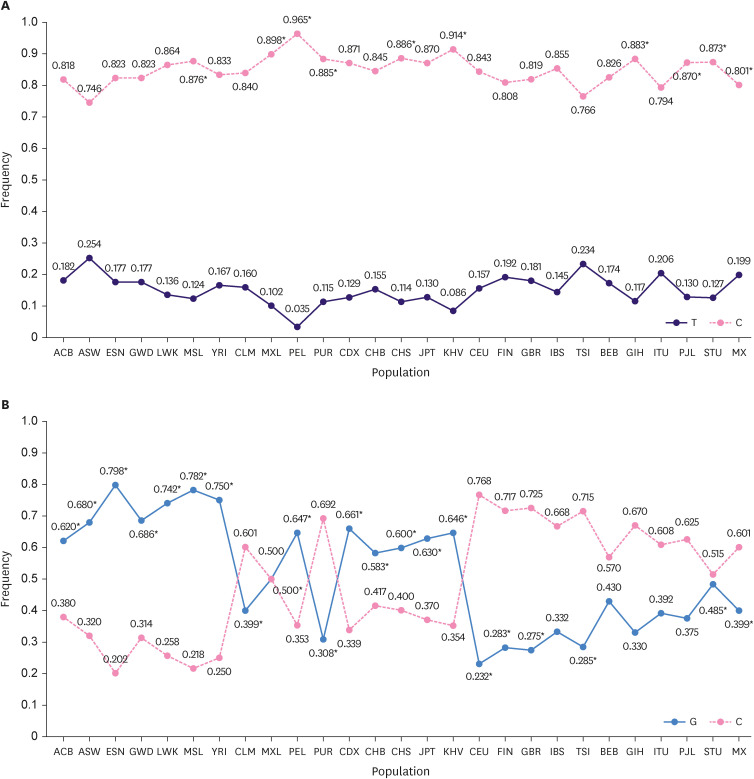
Comparative analysis of rs1047972 and rs8173 variants of the AURKA gene in the control group of Mexican women in this study with a control group of other populations showed statistically significant differences in allelic distribution (Table 3, Figure 1).
Table 3. Allelic distribution of the rs1047972, and rs8173 variants of AURKA in controls from different populations.
| Population | rs1047972 | rs8173 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele C | No. | Allele T | No. | p-value | Allele C | No. | Allele G | No. | p-value | |
| ACB | 0.818 | 157 | 0.182 | 35 | 0.70 | 0.380 | 73 | 0.620 | 119 | 0.0001† |
| ASW | 0.746 | 91 | 0.254 | 31 | 0.21 | 0.320 | 39 | 0.680 | 83 | 0.0001† |
| ESN | 0.823 | 186 | 0.177 | 35 | 0.23 | 0.202 | 40 | 0.798 | 158 | 0.0001† |
| GWD | 0.823 | 186 | 0.177 | 40 | 0.55 | 0.314 | 71 | 0.686 | 155 | 0.0001† |
| LWK | 0.864 | 171 | 0.136 | 27 | 0.06 | 258 | 51 | 0.742 | 147 | 0.0003† |
| MSL | 0.876 | 149 | 0.124 | 21 | 0.035† | 218 | 37 | 0.782 | 133 | 0.0001† |
| YRL | 0.833 | 180 | 0.167 | 36 | 0.36 | 0.250 | 54 | 0.750 | 162 | 0.0001† |
| CLM | 0.840 | 158 | 0.160 | 30 | 0.28 | 0.601 | 113 | 0.399 | 75 | 0.0001† |
| MXL | 0.898 | 115 | 0.102 | 13 | 0.01† | 0.500 | 64 | 0.500 | 64 | 0.038† |
| PEL | 0.965 | 164 | 0.035 | 6 | 0.0001† | 0.353 | 60 | 0.647 | 110 | 0.0001† |
| PUR | 0.885 | 184 | 0.115 | 24 | 0.01† | 0.692 | 144 | 0.308 | 64 | 0.01† |
| CDX | 0.871 | 162 | 0.129 | 24 | 0.04† | 0.339 | 63 | 0.661 | 123 | 0.0001† |
| CHB | 0.845 | 174 | 0.155 | 32 | 0.21 | 0.417 | 86 | 0.583 | 120 | 0.0001† |
| CHS | 0.886 | 186 | 0.114 | 24 | 0.008† | 0.400 | 84 | 0.600 | 126 | 0.0001† |
| JPT | 0.870 | 181 | 0.130 | 27 | 0.03† | 0.370 | 77 | 0.630 | 131 | 0.0001† |
| KHV | 0.914 | 181 | 0.086 | 17 | 0.0004† | 0.354 | 70 | 0.646 | 129 | 0.0001† |
| CEU | 0.843 | 167 | 0.157 | 31 | 0.23 | 0.768 | 152 | 0.232 | 46 | 0.0001† |
| FIN | 0.808 | 160 | 0.192 | 38 | 0.92 | 0.717 | 142 | 0.283 | 56 | 0.003† |
| GBR | 0.819 | 149 | 0.181 | 33 | 0.68 | 0.725 | 132 | 0.275 | 50 | 0.002† |
| IBS | 0.855 | 183 | 0.145 | 31 | 0.10 | 0.668 | 143 | 0.332 | 71 | 0.06 |
| TSI | 0.766 | 164 | 0.234 | 50 | 0.33 | 0.715 | 153 | 0.285 | 61 | 0.002† |
| BEB | 0.826 | 142 | 0.174 | 30 | 0.55 | 0.570 | 98 | 0.430 | 74 | 0.49 |
| GIH | 0.883 | 182 | 0.117 | 24 | 0.01† | 0.670 | 138 | 0.330 | 68 | 0.08 |
| ITU | 0.794 | 162 | 0.206 | 42 | 0.90 | 0.608 | 124 | 0.392 | 80 | 0.93 |
| PJL | 0.870 | 167 | 0.130 | 25 | 0.044† | 0.625 | 120 | 0.375 | 72 | 0.60 |
| STU | 0.873 | 178 | 0.127 | 26 | 0.03† | 0.515 | 105 | 0.485 | 99 | 0.03† |
| MX* | 0.801 | 436 | 0.199 | 108 | 0.601 | 492 | 0.399 | 346 | ||
ACB = African Caribbean in Barbados, by its acronym in Spanish; ASW = African Caribbean in Barbados; ESN = Esan in Nigeria; GWD = Gambian in Western Division; LWK = Luhya in Webuye, Kenya; MSL = Mende in Sierra Leone; YRI = Yoruba in Ibadan, Nigeria; CLM = Colombian in Medellin; MXL = Mexican Ancestry in Los Angeles, California; PEL = Peruvian in Lima; PUR = Puerto Rican in Puerto Rico; CDX = Chinese Dai in Xishuangbanna; CHB = Han Chinese in Beijing; CHS, = Southern Han Chinese; JPT = Japanese in Tokyo; KHV = Kinh in Ho Chi Minh City, Vietnam; CEU = Utah residents with Northern and Western European ancestry; FIN = Finnish in Finland; GBR = British in England and Scotland; IBS = Iberian Populations in Spain; TSI = Tuscan in Italy; BEB = Bengali in Bangladesh; GIH = Gujarati Indian in Houston; ITU = Indian Telugu in the UK; PJL = Punjabi in Lahore, Pakistan; STU = Sri Lankan Tamil in the UK; MX = Mexican population.
*MX studied the T and G alleles of rs1047972, and rs8173 variants of AURKA respectively, were too compared with other population.
†p < 0.05. The frequencies were taken from Ensembl CRCh38.p13.
Figure 1. Allelic frequency comparison of the rs1047972 (A) and rs8173 (B) variants of the AURKA gene in the Mexican women control group with controls from other populations.
When comparing the frequency of the T and C alleles of the rs1047972 and C and G alleles of the rs8173 variant in the Mexican population with those of other populations, significant differences were observed (p < 0.05).
ACB = African Caribbean in Barbados; ASW = African Ancestry in Southwest US; ESN = Esan in Nigeria; GWD = Gambian in Western Division, The Gambia; MSL = Mende in Sierra Leone; YRI = Yoruba in Ibadan, Nigeria; CLM = Colombian in Medellin, Colombia; MXL = Mexican Ancestry in Los Angles, California; PEL = Peruvian in Lima, Peru; PUR = Puerto Rican in Puerto Rico; CDX = Chinese Dai in Xishuangbanna, China; CHB = Han Chinese in Beijing, China; CHS = Southern Han Chinese, China; JPT = Japanese in Tokyo, Japan; KHV = Kinh in Ho Chi Minh City, Vietnam; CEU = Utah resident with Northern and Western European ancestry; GBR = British in England and Scotland; BEB = Bengali in Bangladesh; ITU = Indian Telugu in the UK; STU = Sri Lankan Tamil in the UK; MX = Mexican population from our study.
*Data were presented in Table 3 and in the text and the frequencies were taken from Ensembl database using the reference genome “CRCh38.p13.”
No statistically significant differences were observed for either rs1047972 or rs8173 variants regarding the variables age (stratified in ≥ 50 years and ≤ 49 years), and tobacco and alcohol consumption between the BC and control groups.
In the BC group, the CT genotype (OR, 2.6; 95% CI, 1.26–5.67; p = 0.010) and the dominant model (OR, 2.3; 95% CI, 1.17–4.46; p = 0.016) of the rs1047972 variant were associated with overweight and obesity (CT genotype: OR, 3.2; 95% CI, 1.60–6.66; p = 0.001 and dominant model: OR, 2.5; 95% CI, 1.32–4.87; p = 0.005) as risk factors for BC susceptibility. Likewise, the dominant model (OR, 1.9; 95% CI, 1.03–3.68; p = 0.038) showed an association of susceptibility to BC risk with those patients who had a family history of cancer (Table 4).
Table 4. rs1047972 and rs8173 variants of the AURKA gene and their association with the breast cancer group.
| Variant | Genotype | Variable | OR (95% CI) | p-value |
|---|---|---|---|---|
| rs1047972 | CT | Overweight | 2.6 (1.26–5.67) | 0.010 |
| CTTT | 2.3 (1.17–4.46) | 0.016 | ||
| CT | Obesity | 3.2 (1.60–6.66) | 0.001 | |
| CTTT | 2.5 (1.32–4.87) | 0.005 | ||
| CTTT | Family history of cancer | 1.9 (1.03–3.68) | 0.038 | |
| CTTT | Estrogen receptor plus obesity | 2.5 (1.11–5.79) | 0.027 | |
| CTTT | Ki-67 (≥ 20%) plus family history of cancer | 2.66 (1.14–6.2) | 0.023 | |
| rs8173 | CG | Chemotherapy gastric toxicity | 1.6 (1.05–2.46) | 0.029 |
| CGGG | 1.7 (1.11–2.72) | 0.015 | ||
| CGGG | Estrogen receptor plus chemotherapy gastric toxicity | 2.0 (1.15–3.48) | 0.013 | |
| CGGG | Progesterone receptor plus chemotherapy gastric toxicity | 2.6 (1.42–4.9) | 0.002 | |
| CGGG | Menopause plus chemotherapy gastric toxicity | 2.0 (1.15–3.7) | 0.014 |
p-value less than 0.05 considered statistically significant.
OR = odds ratio; CI =confidence intervals.
Regarding the rs8173 variant, the CG genotype (OR, 1.6; 95% CI, 1.05–2.46; p = 0.029), and dominant model (OR, 1.7; 95% CI, 1.11–2.72; p = 0.015) were associated with chemotherapy gastric toxicity as BC susceptibility risk factors (Table 4).
Patients with BC who were carriers of the CTTT genotype of the rs1047972 variant with elevated Ki-67 and who have a familial antecedent with cancer (OR, 2.66; 95% CI, 1.14–6.2; p = 0.023), showed risk factor of the susceptibility of BC (Table 4).
The haplotype and frequency comparisons among the studied group were significantly different in CG (OR, 0.7; 95% CI, 0.53–0.93; p = 0.014), and TC (OR, 1.6; 95% CI, 1.11–2.32; p = 0.010) (Table 5). The linkage disequilibrium of the rs1047972 and rs8173 variants was D′ 0.072 and r′ = 0.003, in the control group.
Table 5. Haplotype frequency of the rs1047972 and rs8173 variants of the AURKA gene in studied groups.
| rs1047972 | rs8173 | BC (2n = 548) | Controls (2n = 446) | OR (95% CI) | p-value |
|---|---|---|---|---|---|
| C | C | 259 (47.3) | 218 (48.9) | 0.9 (0.73–1.20) | 0.629 |
| C | G | 130 (23.7) | 138 (30.9) | 0.7 (0.53–0.93) | 0.014 |
| T | C | 93 (17.0) | 51 (11.4) | 1.6 (1.11–2.32) | 0.010 |
| T | G | 66 (12.0) | 41 (9.2) | 1.3 (0.88–2.02) | 0.162 |
All those frequencies < 0.03 were not taken into account for the analysis. Values are presented as number (%) not otherwise specified.
BC = breast cancer; OR = odds ratio; CI = confidence interval.
DISCUSSION
BC is a major health concern worldwide. In Mexico, it is the main cause of death in women, and its incidence has been increasing in the last 10 years, with the highest frequency observed in women aged approximately 50 years. This finding is consistent with the average age data observed in the present study. However, in developed and developing countries such as Mexico, the presence of reproductive and hormonal risk factors and those related to lifestyle reflects the increase in the incidence of BC [1,2,30]. Among the clinical features observed as a prevalent pattern in the samples analyzed in this study were menopause, obesity, less than five pregnancies, location of unilateral cancer, ductal type, advanced stages II and III, positive lymphoma nodules, and luminal A/B type of cancer. A characteristic fact It should be highlighted that most patients are in the advanced stages of the disease.
AURKA is a protein that participates in the formation of microtubules and stabilization of chromosomes during the cell cycle; its expression levels are high in the lung, breast, and testis, which makes it an important molecule in the microenvironment and proliferation of this type of tumor. Different studies in the literature have described contradictory results, some have described the association of the rs1047972 (Aurora-A V57I) variant with reduced risk of developing cancer, others with susceptibility to the risk of developing cancer, and others have not described an association [12,18,21,22,23,24,25,26,27,28].
In the present study, the TT and CTTT (dominant model) genotypes and the T allele of variant rs1047972 were associated with the risk of developing BC (p < 0.05). This is the first study carried out in a Mexican population where this association of susceptibility to the risk of developing BC is evidenced. Previous studies carried out on BC in other populations have found a protective association, when it comes to case-control studies [19,21,22]. However, a study conducted on patients with BC and mutations in the BRCA1 gene in a Chinese population observed an association between cancer risk and variants of the AURKA gene [31].
In the rs8173 variant, we observed that the GG genotype and the recessive model were associated with protective factors against the development of BC (p < 0.05). There are no association studies with BC and a control group; however, our data are consistent with a study conducted on children with Wilms tumors in a Chinese population [32].
When comparing the T and G alleles of rs1047972 and rs8173, respectively, of our Mexican women control group with the control groups of other populations, similarities and differences were observed in Asian, African, Latin, and Caucasian populations. These results show the genetic heterogeneity of these variants in other populations, and it is important to highlight the importance of the sample size used in different population analyses [30,33].
We observed that the CT and CTTT carriers of the rs1047972 variant and CG and CGGG carriers of the rs8173 variant were risk factors for the development of BC, as stratified by different clinicopathological parameters, such as overweight, obesity, family history of cancer, chemotherapy gastric toxicity, estrogen receptor-positive plus obesity, Ki-67 > 20% plus family history of cancer, and estrogen receptor-positive plus chemotherapy gastric toxicity. Although no existing studies support these findings, these confounding factors show that stratification is important in contributing to the differences in rs1047972 and rs8173 variants and their associations with BC risk. However, an association study with variants of the AURKA gene and clinical pathological characteristics of BC in patients from Swedish and Polish families showed that the G allele of the rs8173 variant of the AURKA gene was associated with steroid hormone receptor status; moreover, they suggested that this same allele may be associated with worse survival in patients with BC in hormone-dependent tumors [34].
AURKA overexpression has been shown to be essential for tumor survival, and its activity is modulated by various recognition sites that bind AURKA to regulate its expression such as epidermal growth factor receptor, signal transducer and activator of transcription 5, Ajuba, forkhead box M1, targeting protein for Xklp2, and pumilio RNA-binding family member 2 [12]. Furthermore, AURKA protein participates in the modulation of various cell regulation pathways, such as kinase cascades, growth factors, Kirsten rat sarcoma viral oncogene homolog, regulation of centromeres, cell cycle, binding of estrogen receptor α to DNA, and promotion of cell proliferation in cancer and chemoresistance. Probably, the variant alleles of rs1047972 (T) and rs8173 (G) of AURKA are located at the target sites of cell recognition and modulation, and as a consequence, may influence the imbalance of AURKA and the survival of cancer cells [12].
Our study investigated the rs8173 (C>G) variant and found that the GG genotype was a protective factor, consistent with previous findings. Specifically, normal tissues have shown that the CC genotype has a statistically significant difference in median expression compared to the GG genotype in cultured fibroblasts [32]. In addition, another study reported that the rs2273535 variant, which has an effect similar to that of rs1047972, is associated with poor prognosis in gastric cancer in the Chinese population. According to their bioinformatics analysis, the variant genotype could lead to higher expression of the AURKA gene, which is also associated with poor prognosis of the disease [35].
Regarding the study limitations and concerning the sample size, a larger population sample might aid in adjusting our statistical findings. Additionally, no molecular screening for cancer-related pathogenic variants (such as BRCA1/2) was carried out in our study groups. Thus, excluding these subjects would generate a better fit for our statistical analyses.
The AURKA variants analyzed in this study were not in linkage disequilibrium. Two frequent haplotypes rs1047972 and rs8173 were observed in the study groups analyzed. The CG haplotype, present in 31% of controls and 24% of patients with BC, is a protective factor for the development of BC. Haplotype TC, present in 11% of the control group and 17% of the patient group, was associated with susceptibility to BC. Unfortunately, no studies on BC have analyzed this association. Thus, the combination of the two variants of AURKA analyzed in this study may confer a significant association with risk predisposition for BC, especially with the rs1047972 and rs8173 variants.
Our results showed characteristic phenotypes in patients with BC: histological ductal type, luminal types A/B, lymph node metastasis, and advanced stages III–IV. The rs1047972 variant was found to be an associated risk factor for BC when controls and patients with BC with the genotype TT, allele T, and the dominant model (CTTT genotype) were compared. The rs8173 variant was found to be a protective factor against BC when the groups with the GG genotype were compared.
In addition, we observed statistically significant differences in the rs1047972 variant showing a risk association when the group of patients with BC was stratified by the CT genotype and the dominant model (CTTT), with overweight and obesity, and the presence of a history of familial antecedents of cancer, estrogen receptor marker positivity plus obesity, and marker Ki-67 > 20% plus a family history of cancer.
The rs8173 variant was also identified as a risk factor when the BC patient group carrier of the CT genotype was stratified by chemotherapy gastric toxicity, model-dominant CTTT with hormonal receptor plus chemotherapy gastric toxicity, and chemotherapy gastric toxicity plus menopausal status. The presence of the TC haplotype is a risk factor for BC. However, further studies are needed to confirm these findings.
As a future perspective, the inclusion of more variants related to the AURKA gene, quantification of the expression levels, and their correlation with other related genes might be helpful in further association studies in our population.
Footnotes
Conflict of Interest: The authors declare that they have no competing interests.
- Conceptualization: Maciel-Cruz EJ, Figuera Villanueva LE, Garibaldi-Ríos AF, Zúñiga-Gónzalez GM, Gallegos-Arreola MP.
- Data curation: Maciel Cruz EJ, Figuera Villanueva LE, Garibaldi-Ríos AF, Zúñiga-Gónzalez GM, Gallegos-Arreola MP.
- Formal analysis: Maciel Cruz EJ, Garibaldi-Ríos AF, Gómez-Meda BC, Zúñiga-Gónzalez GM, Pérez AM, Castro-García PB, Ramírez-Patiño R, Gallegos-Arreola MP.
- Investigation: Figuera Villanueva LE.
- Methodology: Maciel Cruz EJ, Garibaldi-Ríos AF, Gómez-Meda BC, Zúñiga-Gónzalez GM, Pérez AM, Castro-García PB, Ramírez-Patiño R, Gallegos-Arreola MP.
- Writing - original draft: Maciel Cruz EJ, Figuera Villanueva LE, Garibaldi-Ríos AF, Zúñiga-Gónzalez GM, Gallegos-Arreola MP.
- Writing - review & editing: Maciel Cruz EJ, Figuera Villanueva LE, Garibaldi-Ríos AF, Zúñiga-Gónzalez GM, Gallegos-Arreola MP.
References
- 1.Kashyap D, Pal D, Sharma R, Garg VK, Goel N, Koundal D, et al. Global increase in breast cancer incidence: risk factors and preventive measures. BioMed Res Int. 2022;2022:9605439. doi: 10.1155/2022/9605439. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol. 2022;95:20211033. doi: 10.1259/bjr.20211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trapani D, Ginsburg O, Fadelu T, Lin NU, Hassett M, Ilbawi AM, et al. Global challenges and policy solutions in breast cancer control. Cancer Treat Rev. 2022;104:102339. doi: 10.1016/j.ctrv.2022.102339. [DOI] [PubMed] [Google Scholar]
- 4.Picazo J, Lassard Rosenthal J, Aguilar L, Núñez C. Breast cancer: an overview. Acta Med Grupo Ángeles. 2021;19:354–360. [Google Scholar]
- 5.Sarhangi N, Hajjari S, Heydari SF, Ganjizadeh M, Rouhollah F, Hasanzad M. Breast cancer in the era of precision medicine. Mol Biol Rep. 2022;49:10023–10037. doi: 10.1007/s11033-022-07571-2. [DOI] [PubMed] [Google Scholar]
- 6.Rooney MM, Miller KN, Plichta JK. Genetics of breast cancer: risk models, who to test, and management options. Surg Clin North Am. 2023;103:35–47. doi: 10.1016/j.suc.2022.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Yiangou K, Kyriacou K, Kakouri E, Marcou Y, Panayiotidis MI, Loizidou MA, et al. Combination of a 15-SNP polygenic risk score and classical risk factors for the prediction of breast cancer risk in Cypriot women. Cancers (Basel) 2021;13:4568. doi: 10.3390/cancers13184568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Fidalgo JA, Gambardella V, Pineda B, Burgues O, Piñero O, Cervantes A. Aurora kinases in ovarian cancer. ESMO Open. 2020;5:e000718. doi: 10.1136/esmoopen-2020-000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan M, Wang C, He B, Yang M, Tong M, Long Z, et al. Aurora-A kinase: a potent oncogene and target for cancer therapy. Med Res Rev. 2016;36:1036–1079. doi: 10.1002/med.21399. [DOI] [PubMed] [Google Scholar]
- 10.Shan B, Zhao R, Zhou J, Zhang M, Qi X, Wang T, et al. AURKA increase the chemosensitivity of colon cancer cells to oxaliplatin by inhibiting the TP53-mediated DNA damage response genes. BioMed Res Int. 2020;2020:8916729. doi: 10.1155/2020/8916729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiwari S, Pandey VP, Yadav K, Dwivedi UN. Modulation of interaction of BRCA1-RAD51 and BRCA1-AURKA protein complexes by natural metabolites using as possible therapeutic intervention toward cardiotoxic effects of cancer drugs: an in-silico approach. J Biomol Struct Dyn. 2022;40:12863–12879. doi: 10.1080/07391102.2021.1976278. [DOI] [PubMed] [Google Scholar]
- 12.Du R, Huang C, Liu K, Li X, Dong Z. Targeting AURKA in cancer: molecular mechanisms and opportunities for cancer therapy. Mol Cancer. 2021;20:15. doi: 10.1186/s12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miralaei N, Majd A, Ghaedi K, Peymani M, Safaei M. Integrated pan-cancer of AURKA expression and drug sensitivity analysis reveals increased expression of AURKA is responsible for drug resistance. Cancer Med. 2021;10:6428–6441. doi: 10.1002/cam4.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, Zhang Y, Dong Y, Ning P, Zhang Y, Sun H, et al. Knockdown of AURKA sensitizes the efficacy of radiation in human colorectal cancer. Life Sci. 2021;271:119148. doi: 10.1016/j.lfs.2021.119148. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Qi J, Zhu M, Wang M, Nie J. AURKA rs2273535 T>A polymorphism associated with cancer risk: a systematic review with meta-analysis. Front Oncol. 2020;10:1040. doi: 10.3389/fonc.2020.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Chen L, Zhang Y, Wang L, Zheng W, Peng F, et al. Predicting AURKA as a novel therapeutic target for NPC: a comprehensive analysis based on bioinformatics and validation. Front Genet. 2022;13:926546. doi: 10.3389/fgene.2022.926546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CH, Chen CJ, Chen PN, Wang SS, Chou YE, Hung SC, et al. Impacts of AURKA genetic polymorphism on urothelial cell carcinoma development. J Cancer. 2019;10:1370–1374. doi: 10.7150/jca.30014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farid AA, Afify NA, Alsharnoby AA, Abdelsameea E, Bedair HM. Predictive role of AURKA rs1047972 gene polymorphism and the risk of development of hepatocellular carcinoma. Immunol Invest. 2022;51:1211–1221. doi: 10.1080/08820139.2021.1920609. [DOI] [PubMed] [Google Scholar]
- 19.Wu PJ, Wang CH, Hsieh MH, Lee CY, Wang PH, Lin CW, et al. The impact of Aurora kinase A genetic polymorphisms on cervical cancer progression and clinicopathologic characteristics. Int J Med Sci. 2021;18:2457–2465. doi: 10.7150/ijms.58516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W, Qiu H, Jiang H, Wang L, Sun B, Gu H. Aurora-A V57I (rs1047972) polymorphism and cancer susceptibility: a meta-analysis involving 27,269 subjects. PLoS One. 2014;9:e90328. doi: 10.1371/journal.pone.0090328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai ZJ, Kang HF, Wang XJ, Shao YP, Lin S, Zhao Y, et al. Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer Cell Int. 2014;14:91. doi: 10.1186/s12935-014-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor NJ, Bensen JT, Poole C, Troester MA, Gammon MD, Luo J, et al. Genetic variation in cell cycle regulatory gene AURKA and association with intrinsic breast cancer subtype. Mol Carcinog. 2015;54:1668–1677. doi: 10.1002/mc.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Hu C, Song Y, Xiu M, Zhang Y, Lai P, et al. Relationship between Aurora-A V57I polymorphism and the risk of cancer: a meta-analysis and trial sequential analysis. J Cancer. 2020;11:3225–3234. doi: 10.7150/jca.40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesic A, Rogar M, Hudler P, Juvan R, Komel R. Association of the AURKA and AURKC gene polymorphisms with an increased risk of gastric cancer. IUBMB Life. 2016;68:634–644. doi: 10.1002/iub.1521. [DOI] [PubMed] [Google Scholar]
- 25.Tang J, Qian Y, Zhu J, Zhang J, Wang FH, Zeng JH, et al. Lack of associations between AURKA gene polymorphisms and neuroblastoma susceptibility in Chinese children. Biosci Rep. 2018;38:BSR20180292. doi: 10.1042/BSR20180292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang X, Zhang H, Lian S, Zhu W. miR-137 suppresses tumor growth of malignant melanoma by targeting aurora kinase A. Biochem Biophys Res Commun. 2016;475:251–256. doi: 10.1016/j.bbrc.2016.05.090. [DOI] [PubMed] [Google Scholar]
- 27.Xu L, Liu H, Yan Z, Sun Z, Luo S, Lu Q. Inhibition of the Hedgehog signaling pathway suppresses cell proliferation by regulating the Gli2/miR-124/AURKA axis in human glioma cells. Int J Oncol. 2017;50:1868–1878. doi: 10.3892/ijo.2017.3946. [DOI] [PubMed] [Google Scholar]
- 28.Rykova E, Ershov N, Damarov I, Merkulova T. SNPs in 3'UTR miRNA target sequences associated with individual drug susceptibility. Int J Mol Sci. 2022;23:13725. doi: 10.3390/ijms232213725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 30.Gallegos-Arreola MP, Ramírez-Patiño R, Sánchez-López JY, Zúñiga-González GM, Figuera LE, Delgado-Saucedo JI, et al. SOD2 gene variants (rs4880 and rs5746136) and their association with breast cancer risk. Curr Issues Mol Biol. 2022;44:5221–5233. doi: 10.3390/cimb44110355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruan Y, Song AP, Wang H, Xie YT, Han JY, Sajdik C, et al. Genetic polymorphisms in AURKA and BRCA1 are associated with breast cancer susceptibility in a Chinese Han population. J Pathol. 2011;225:535–543. doi: 10.1002/path.2902. [DOI] [PubMed] [Google Scholar]
- 32.Lu T, Li L, Zhu J, Liu J, Lin A, Fu W, et al. AURKA rs8173 G>C polymorphism decreases Wilms tumor risk in Chinese children. J Oncol. 2019;2019:9074908. doi: 10.1155/2019/9074908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallegos-Arreola MP, Zúñiga-González GM, Figuera LE, Puebla-Pérez AM, Márquez-Rosales MG, Gómez-Meda BC, et al. ESR2 gene variants (rs1256049, rs4986938, and rs1256030) and their association with breast cancer risk. PeerJ. 2022;10:e13379. doi: 10.7717/peerj.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi H, Bevier M, Johansson R, Grzybowska E, Chen B, Eyfjörd JE, et al. Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast Cancer Res Treat. 2011;130:905–916. doi: 10.1007/s10549-011-1600-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Wang P, Zhao H. The association between AURKA gene rs2273535 polymorphism and gastric cancer risk in a Chinese population. Front Physiol. 2018;9:1124. doi: 10.3389/fphys.2018.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]