Figure 2.
Combining datasets to predict values and uncertainties for missing viruses
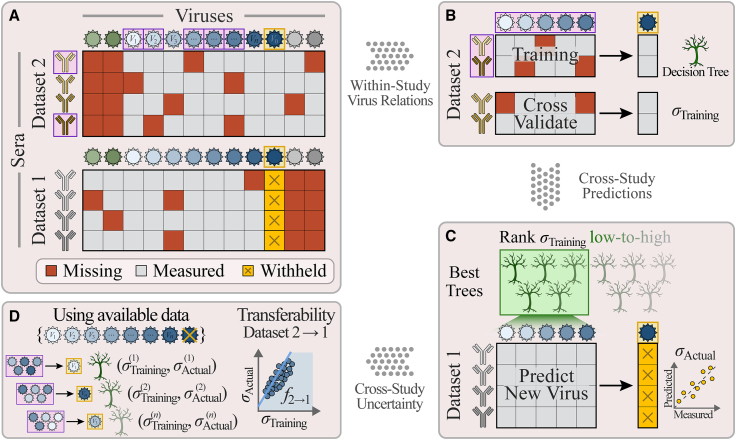
(A) Schematic of data availability; two studies measure antibody responses against overlapping viruses (shades of blue) as well as unique viruses (green/gray). Studies may have different fractions of missing values (dark-red boxes) and measured values (gray). To test whether virus behavior can be inferred across studies, we predict the titers of a virus in dataset 1 (V0, gold squares), using measurements from the overlapping viruses (V1–Vn) as features in a random forest model.
(B) We train a decision tree model using a random subset of antibodies and viruses from dataset 2 (boxed in purple), cross-validate against the remaining antibody responses in dataset 2, and compute the root-mean-square error (RMSE, denoted by σTraining).
(C) Multiple decision trees are trained, and the average from the 5 trees with the lowest error are used as the model going forward. Applying this model to dataset 1 (which was not used during training) yields the desired predictions, whose RMSE is given by σActual. We repeat this process, withholding each virus in every dataset.
(D) To estimate the prediction error σActual (which we are not allowed to directly compute because V0’s titers are withheld), we define the transferability relation f2→1 between the training error σTraining in dataset 2 and actual error σActual in dataset 1 using the decision trees that predict viruses V1–Vn (without using V0). Applying this relation to the training error, f2→1(σTraining), estimates σActual for V0.