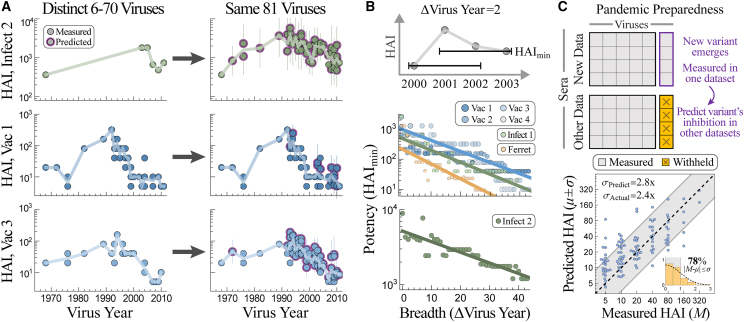
Figure 7.
Applications of cross-study predictions
(A) We predict HAI titers for 25,000 sera against the same set of 81 viruses, providing high-resolution landscapes that can be directly compared against each other. Representative responses are shown for datasetInfect,2 (top, serum 5130165 in GitHub), datasetVac,1 (center, subject 525), and datasetVac,3 (bottom, subject A028).
(B) Tradeoff between serum breadth and potency, showing that viruses spaced apart in time are harder to simultaneously inhibit. For every study and each possible set of viruses circulating within Δvirus years of each other, we calculate the highest HAImin (i.e., a serum exists with HAI titers ≥ HAImin against the entire set of viruses).
(C) Top: when a new variant emerges and is measured in a single study, we can predict its titers in all previous studies with ≥5 overlapping viruses. Bottom: example predicting how the newest variant in the newest vaccine dataset is inhibited by sera from a previous vaccine study (datasetsVac,4→Vac,3).