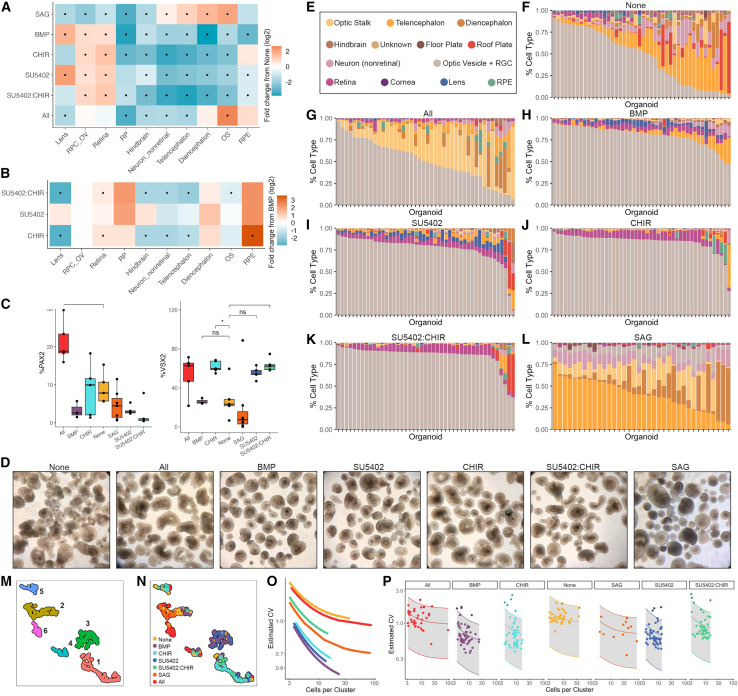
Figure 3.
sci-Plex allows the detection of significant changes in cell-type abundance and heterogeneity
(A) Heatmap of fold change in cell-type abundance compared to “None” for day 28 organoids. The data was modeled using a beta-binomial distribution and significance was determined by a Wald test. An asterisk (∗) in the center of the box indicates a q value of less than 0.05 after Benjamini-Hochberg correction.
(B) Heatmap of fold change in cell-type abundance compared with BMP for day 28 organoids. Statistical analysis was performed as in (A).
(C) Quantification of day 27 immunostained organoids. Significance was determined by performing t tests between the treatment of interest and None. Benjamini-Hochberg correction was performed to adjust the p value. ∗p < 0.05. The line of the boxplots represents the median cells per organoid. The colored region indicates the values from the lower 25th to the upper 75th quartile of cell counts. The whiskers span the range of the cell counts within 1.5× the distance between the 25th–75th percentiles.
(D) Bright-phase images of organoids the day before sci-Plex (day 27). Scale bars represent 200 μM.
(E) Cell-type legend for (F)–(L).
(F) Stacked bar plot, each bar represents an individual day 28 organoid exposed to the None treatment. Bars colored by cell-class composition.
(G–L) Same as (F) but for (G) All, (H) BMP, (I) SU5402, (J) CHIR, (K) SU5402:CHIR, and (L) SAG.
(M) UMAP generated using the size-factor-normalized cell type by organoid matrix generated from day 28 individual organoids. Colored by cluster.
(N) UMAP from (M) with the organoids colored by treatment.
(O) The mean and coefficient of variation (CV) were modeled for each cluster and each treatment. Displayed are the modeled relationships colored by treatment.
(P) Model from (O) faceted by treatment. The shaded area marks the range within 2 standard deviations from the mean. Points represent the individual cluster values.