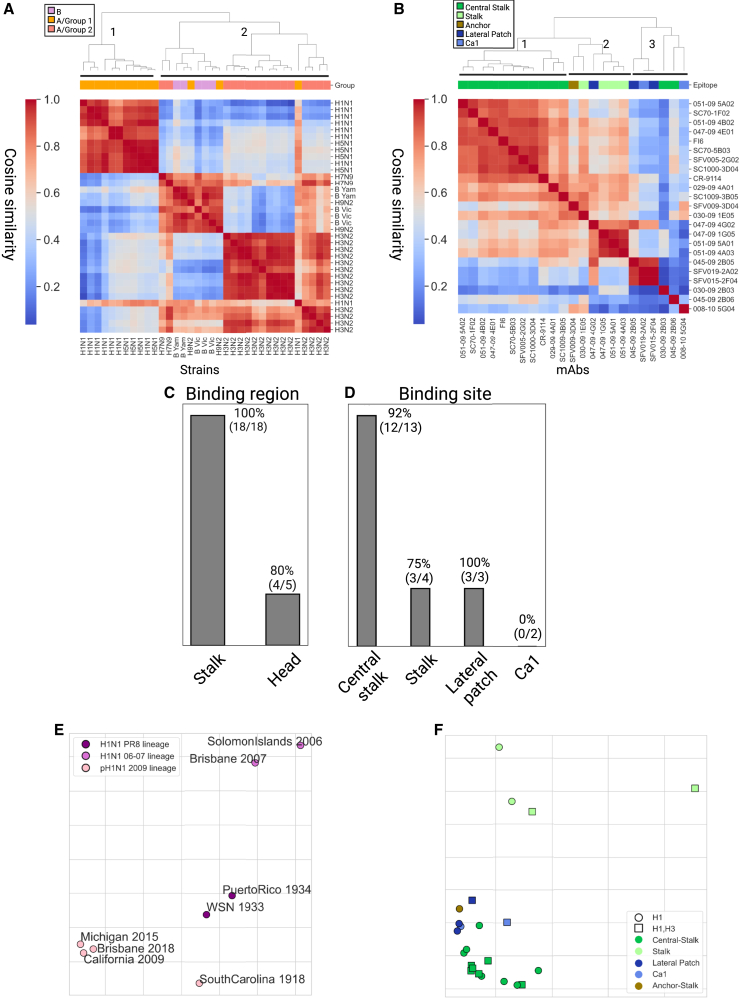
Figure 3.
Inferring antigenic cartography based on mAb binding antibody profiles
(A and B) The mAb binding profiles across HA strains were used to compute pairwise similarities between influenza strains and monoclonal antibodies using cosine similarity. Dendrograms were computed using the complete-linkage algorithm.
(A) Pairwise cosine similarity matrix of 35 influenza HAs from different subtypes computed over the binding profile of all 23 mAbs to each strain. The top color bar represents the antigenic group of each strain (group 1: H1N1, H5N1, and H9N2; group 2: H3N2 and H7N9).
(B) Pairwise cosine similarity matrix of the 23 mAbs computed using the binding profiles to 35 HA strains (Table S1).
(C and D) Classification accuracy of KNN classification, using k = 3, was used to classify each mAb as follows: (C) binding region (head or stalk) and (D) binding site (central stalk, stalk, lateral patch, and Ca1). A single anchor-stalk-binding mAb was discarded from this analysis.
(E and F) Antigenic cartography maps based on cosine similarity were computed using the Ramacs antigenic cartography package.51
(E) Antigenic cartography of 8 H1N1 strains based on the binding profiles of 23 H1 mAbs. Influenza strains were divided into 3 known antigenic groups, annotated by color: PR8 lineage, SI06 and BRIS07 lineage, and pH1N1 lineage.
(F) Antigenic cartography of 23 influenza mAbs based on their binding profiles to 36 influenza A HA proteins from both human and avian subtypes (Tables 2 and S2). An embedding based on the Euclidean distance metric is provided in Figure S3, and a combined antigenic map of mAbs and antigens is provided in Figure S4. The shape of each mAb represents its known binding profile to H1 and H3 subtypes based on AM binding profiles. The color of each mAb represents its epitope specificity. PR8, Puerto Rico 1934; SI06, Solomon Islands 2006; BRIS07, Brisbane 2007; B Vic, B Victoria lineage; B Yam, B Yamagata lineage.